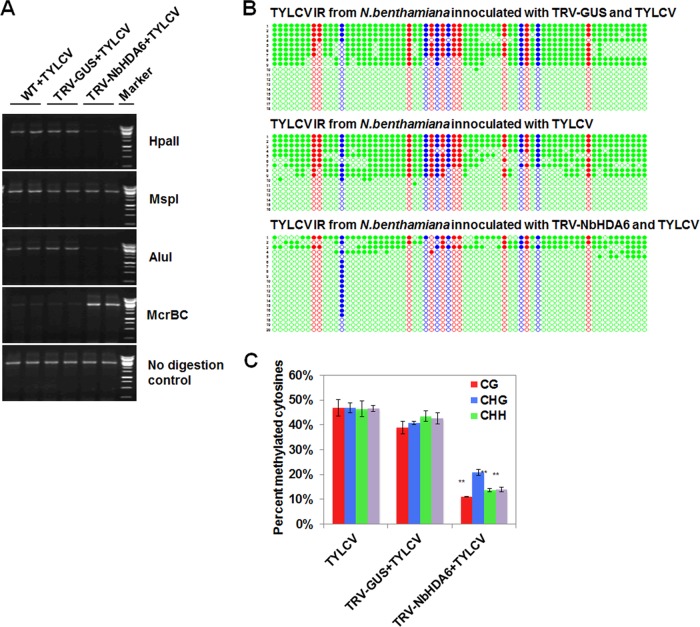
FIG 6.
NbHDA6 positively regulated the cytosine methylation of TYLCV. (A) Analysis of DNA methylation of the TYLCV genome by methylation-sensitive PCR. Genomic DNA was digested with HpaII, MspI, AluI, or McrBC and then loaded into the PCR system. Undigested DNA is shown as a control. (B) Cytosine methylation profiles assessed by bisulfite sequencing. The circles represent cytosine residues and are color coded according to the sequence context (red for CG, blue for CHG, and green for CHH). Solid circles indicate methylated cytosines. Each line represents the sequence of an individual clone. (C) Percentages of methylated cytosines in the TYLCV intergenic regions (IR). Student's t test was performed using the methylation values from individual clones. Double asterisks indicate a significant difference (P < 0.01) between the two-paired samples. Error bars represent the standard errors of the means from independent measurements. Samples were prepared by pooling six leaves from six systemically infected plants at 30 dpi. Similar results were observed in at least three independent experiments.