Cytomegalovirus (CMV) infection is the leading cause of birth defects and causes morbidity and mortality in immunocompromised patients. The regulation of viral late gene transcription is not well elucidated, and understanding of this process benefits the development of novel therapeutics against CMV infection. This study (i) identified that six viral transactivation factors encoded by murine CMV form a complex, (ii) demonstrated that pM91 interacts with pM79 and that pM91 and pM79 colocalize in the nuclear viral replication compartments, (iii) confirmed that pM91 is critical for viral late gene expression but dispensable for viral DNA replication, and (iv) revealed that the pM91-pM79 interaction is required for progeny virus production. These findings give an explanation of how CMV regulates late gene expression and have important implications for the design of antiviral strategies.
KEYWORDS: cytomegalovirus, pM91, pM79, late gene, transcriptional regulation
ABSTRACT
Viral gene expression is tightly regulated during cytomegalovirus (CMV) lytic replication, but the detailed mechanism of late gene transcription remains to be fully understood. Previous studies reported that six viral proteins (named viral transactivation factors [vTFs]) supporting late gene expression were conserved in beta- and gammaherpesviruses but not in alphaherpesviruses. Here, we performed coimmunoprecipitation experiments to elucidate the organization of these six proteins in murine CMV. Our results showed that these proteins formed a complex by both direct and indirect interactions. Specifically, pM91 strongly bound to pM79 even in the absence of other vTFs. Similar to pM79, pM91 exhibited early-late expression kinetics and localized within nuclear viral replication compartments during infection. Functional analysis was also performed using the pM91-deficient virus. Real-time PCR results revealed that abrogation of M91 expression markedly reduced viral late gene expression and progeny virus production without affecting viral DNA synthesis. Using mutagenesis, we found that residues E61, D62, D89, and D96 in pM91 were required for the pM91-pM79 interaction. Disruption of the interaction via E61A/D62A or D89A/D96A double mutation in the context of virus infection inhibited progeny virus production. Our data indicate that pM91 is a component of the viral late gene transcription factor complex and that the pM91-pM79 interaction is essential for viral late gene expression.
IMPORTANCE Cytomegalovirus (CMV) infection is the leading cause of birth defects and causes morbidity and mortality in immunocompromised patients. The regulation of viral late gene transcription is not well elucidated, and understanding of this process benefits the development of novel therapeutics against CMV infection. This study (i) identified that six viral transactivation factors encoded by murine CMV form a complex, (ii) demonstrated that pM91 interacts with pM79 and that pM91 and pM79 colocalize in the nuclear viral replication compartments, (iii) confirmed that pM91 is critical for viral late gene expression but dispensable for viral DNA replication, and (iv) revealed that the pM91-pM79 interaction is required for progeny virus production. These findings give an explanation of how CMV regulates late gene expression and have important implications for the design of antiviral strategies.
INTRODUCTION
Human cytomegalovirus (HCMV), which is also known as human herpesvirus 5, belongs to the Betaherpesvirinae subfamily (1). Following primary infection, HCMV establishes a lifelong latent infection in the host (2). In normal immunocompetent hosts, HCMV infection is usually asymptomatic. However, in immunocompromised hosts, such as transplant recipients, AIDS patients, or neonates, it can cause severe and even life-threatening disease (3, 4). Currently, no licensed vaccine for HCMV is available (5), and the clinical utility of the anti-CMV agents is limited by associated toxicities, poor bioavailability and efficacy, and the risk of resistance with extended use (6–8). Further study is necessary to dissect the roles of viral genes in HCMV infection and will facilitate the effort to develop safer and more effective antiviral therapeutics.
Like all members of the betaherpesviruses, HCMV infection is species specific, and therefore, murine CMV (MCMV) is commonly used in the mouse model to study CMV biology. MCMV shares a series of features with HCMV, including virion structure, genome organization, gene expression program, tissue tropism, and pathogenesis (9, 10). Understanding the functions of conserved viral genes in MCMV will help to analyze the roles of their homologs in HCMV. This may provide novel therapeutic antiviral targets and allow for future testing in mouse models.
Similar to other herpesviruses, CMV lytic replication proceeds through a temporal cascade of gene expression, which can be categorized into three major kinetic classes: immediate early (IE), early, and late (3, 11, 12). IE genes are expressed following viral infection and do not require de novo viral protein synthesis (13). The transcription of early genes requires viral IE proteins but is independent of viral DNA replication (14). Late genes are transcribed following viral DNA synthesis, and their transcription is inhibited by viral DNA synthesis inhibitors, such as phosphonoacetic acid (PAA) (15, 16). These genes mainly encode proteins required for virus assembly and egress (17). In addition, some genes, classified as early-late, are initially expressed prior to viral DNA synthesis but accumulate later in a DNA synthesis-dependent manner. However, although the regulation of IE and early gene transcription has been extensively investigated, the regulation of viral late gene expression is still poorly understood.
Recently, six viral proteins, termed viral transactivation factors (vTFs), have been reported to be essential for late gene expression in gammaherpesviruses (18–24). These vTFs are conserved in beta- and gammaherpesviruses but not in alphaherpesviruses (20, 25). Five, which are expressed in HCMV (pUL79, pUL87, pUL91, pUL92, and pUL95), are found to be involved in regulation of late gene expression (26–29). Specifically, pUL79 participates in RNA polymerase II (RNAP II) elongation (30). The MCMV homologs, pM79 and pM92, are also demonstrated to be key regulators for viral late transcription (16, 31). Recent work in Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) demonstrated that these six vTFs and cellular RNAP II assemble into a viral preinitiation complex (19, 20). Among them, EBV BcRF1 and KSHV ORF24, which are homologs of HCMV pUL87, serve as a TATA box-binding protein (TBP) homolog that binds to RNAP II and TATT sequences in the viral late gene promoter (32–34). In addition, studies in KSHV elucidated clues regarding interactions between vTFs (19, 35, 36). Five homologs (pM49, pM79, pM87, pM92, and pM95) expressed in MCMV were also predicted to form a multicomponent viral complex (37). However, no further experiments were performed to confirm whether the six vTFs in CMV form a complex, and the roles of the vTF complex components require further investigation.
Here, we provide insight into the interactions among the six vTFs expressed in MCMV; we also propose a model of the multicomponent viral complex that regulates late gene transcription. Our findings indicate that four interacting pairs, pM92-pM87, pM87-pM95, pM95-pM49, and pM91-pM79, are involved in forming the complex. In particular, we focused on the pM91-pM79 interaction. The results show that pM91 has an expression pattern similar to that of pM79, localizes with pM79, and is also important for late gene expression but dispensable for viral DNA synthesis. By point mutation analysis, we demonstrate that the pM91-pM79 interaction is essential for infectious virus production. These data suggest that pM91 serves as a component of the viral complex and participates in viral late gene transcription.
RESULTS
Six viral proteins of MCMV form a complex.
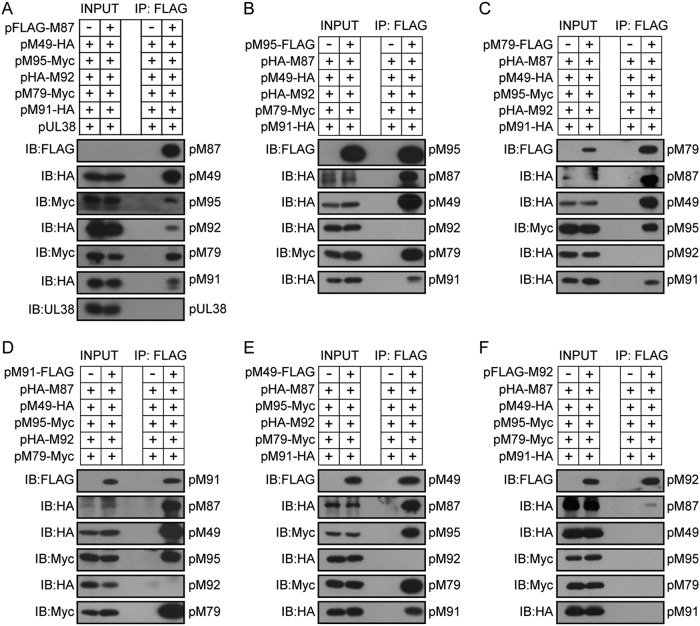
We initially used coimmunoprecipitation assays to confirm whether a MCMV vTF complex similar to that found in gammaherpesviruses exists and to determine its components. HEK293T cells were transfected with the FLAG-tagged pM87 expression plasmid or the empty vector together with the expression plasmids of the other five vTFs. At 48 h posttransfection, the cells were harvested, lysed, and subjected to immunoprecipitation using anti-FLAG M2 (mouse monoclonal) magnetic beads. The elution was analyzed by immunoblotting using the antibodies indicated below. All five vTFs were coimmunoprecipitated in the presence of pM87, while HCMV protein pUL38 was not (Fig. 1A). Similarly, when using either pM95, pM79, pM91, or pM49 as the bait, all other vTFs except pM92 were also coimmunoprecipitated (Fig. 1B to E). Consistent with the results presented above, when pM92 was used as the bait, only a small amount of pM87 was coimmunoprecipitated (Fig. 1F), suggesting that the interaction between pM92 and pM87 was primarily weak. The coimmunoprecipitation results are summarized in Table 1. Together, these results indicate that the six vTFs expressed in MCMV, pM49, pM79, pM87, pM91, pM92, and pM95, are involved in a complex.
FIG 1.
Six MCMV vTFs form a complex. HEK293T cells were cotransfected with the six MCMV vTFs (pM49, pM79, pM87, pM91, pM92, and pM95) with the indicated tags and collected at 48 h posttransfection. HCMV protein pUL38 was used as a negative control. Cell lysates were subjected to immunoprecipitation (IP) using anti-FLAG M2 (mouse monoclonal) magnetic beads, which capture FLAG-tagged pM87 (A), FLAG-tagged pM95 (B), FLAG-tagged pM79 (C), FLAG-tagged pM91 (D), FLAG-tagged pM49 (E), and FLAG-tagged pM92 (F). The whole-cell lysates (INPUT) and the immunoprecipitated fractions were immunoblotted (IB) with the indicated antibodies to detect their interactions.
TABLE 1.
Summary of protein interactions
| Protein | Interaction of the following with the indicated proteins: |
|||||
|---|---|---|---|---|---|---|
| pFLAG-M87 | pM49-FLAG | pM95-FLAG | pFLAG-M92 | pM79-FLAG | pM91-FLAG | |
| pM87 | NTa | ++++ | ++++ | + | ++++ | ++++ |
| pM49 | +++ | NT | ++++ | − | ++++ | ++++ |
| pM95 | + | ++++ | NT | − | +++ | ++++ |
| pM92 | + | − | − | NT | − | − |
| pM79 | ++ | +++ | +++ | − | NT | ++++ |
| pM91 | + | +++ | ++ | − | +++ | NT |
NT, not tested.
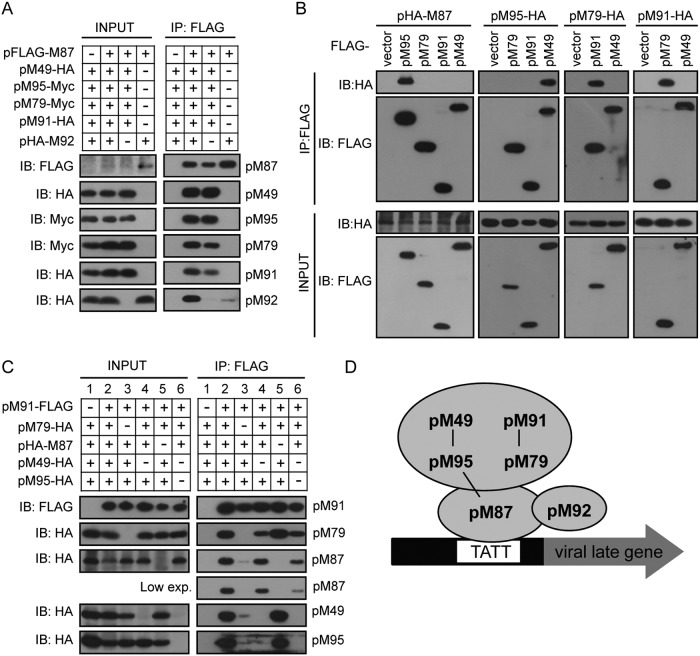
As the affinities of the different vTFs varied (Fig. 1 and Table 1), we hypothesized that the complex probably had a specific organization. To further dissect the organization of the complex, we performed a set of coimmunoprecipitation experiments with different combinations of vTFs in HEK293T cells. As pM92 could be coimmunoprecipitated only with pM87 (Fig. 1F), we first analyzed the interaction between pM87 and pM92: pM87 was able to be coimmunoprecipitated with pM92 alone, but this interaction was greatly enhanced in the presence of the other vTFs (Fig. 2A, lane 2 versus lane 4). Moreover, the amount of the other vTFs coimmunoprecipitated with pM87 was similar in the presence or absence of pM92, which suggests that depletion of pM92 had little effect on the interactions between pM87 and the other vTFs (Fig. 2A, lanes 2 and 3). Thus, we focused on the organization of the viral complex in the absence of pM92.
FIG 2.
Organization of the vTF complex. (A) pM87 interacts with pM92. The FLAG-tagged pM87 expression plasmid or the empty vector was cotransfected with the indicated HA- or Myc-tagged vTFs into HEK293T cells. Cell lysates were immunoprecipitated by anti-FLAG M2 beads and assayed by immunoblotting. (B) Pairwise interaction screening of five vTFs via coimmunoprecipitation. The experiments were performed as described in the legend to panel A, except that the indicated plasmids were used in the samples. (C) Additional interactions were established after coexpression. The experiments were performed as described in the legend to panel A, except that various combinations of vTF subunits, as indicated, were coexpressed in the samples. exp., expression. (D) Model of the MCMV vTF complex. Three pairs of vTF subunits (pM87-pM95, pM95-pM49, and pM91-pM79) form strong interactions, as indicated by black lines. Two of them (pM49-pM95 and pM91-pM79) bind strongly with each other to form a stable subcomplex. The subcomplex binds to pM87 through multiple interfaces. pM92 weakly binds to the complex by interacting with pM87.
To further analyze the organization of the other five vTFs, we performed pairwise interaction screening via coimmunoprecipitation. The results showed three pairs of strong interactions: pM87-pM95, pM95-pM49, and pM91-pM79 (Fig. 2B). As three pairs of interactions did not explain how these six proteins form a complex, we hypothesized that additional interactions could be established after the formation of the protein pairs and we analyzed the interactions between these protein pairs. Although neither pM87, pM95, nor pM49 could be coimmunoprecipitated with pM91 individually (Fig. 2B), they could bind to pM91 weakly when all three were coexpressed (Fig. 2C, lane 3). In addition, this interaction was largely enhanced in the presence of pM79 (Fig. 2C, lane 2). These results suggest that the pM91-pM79 interaction greatly promoted vTF complex formation. When pM49 was absent, the level of pM95 in the immunoprecipitation fraction was greatly reduced and the level of pM87 and pM79 was also relatively decreased (Fig. 2C, lane 4 versus lane 2), suggesting that the pM49-pM95 interaction is also important for vTF complex formation. Moreover, when both pM49 and pM95 were depleted, the level of pM87 in the immunoprecipitation fraction was further decreased (Fig. 2C, lane 6 versus lane 4). It is possible that the formation of pM87-pM95 promoted the interaction between pM91-pM79 and pM87. Interestingly, depletion of pM87 did not impair the level of pM79, pM49, and pM95 in the immunoprecipitation fraction (Fig. 2C, lane 5 versus lane 2), suggesting that pM91-pM79 and pM95-pM49 interacted strongly with each other. A model for the physical interactions between MCMV vTFs is depicted in Fig. 2D.
pM91 shares expression kinetics and localization with pM79.
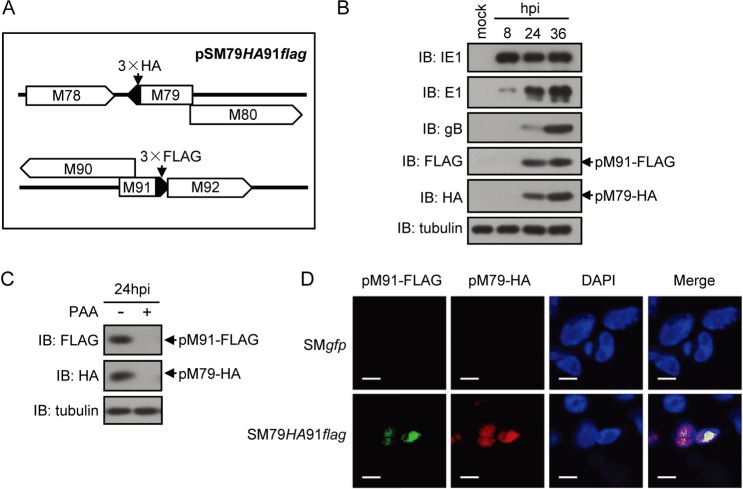
Our results regarding the strong interaction between pM91 and pM79 focused our attention on their functions during infection. pM79 expression was previously reported to be classified as following early-late kinetics, and localization was generally in the nuclear viral replication compartments (16). Therefore, we hypothesized that pM91 had a similar expression pattern and localization. Without available antibodies to pM79 or pM91, we engineered a recombinant virus, SM79HA91flag, by inserting a fragment encoding a 3× hemagglutinin (3×HA) or a 3×FLAG tag in frame at the 3′ end of the M79 or M91 coding sequence, respectively, just upstream of the stop codon (Fig. 3A). We infected mouse embryonic fibroblast 10.1 (MEF10.1) cells with the SM79HA91flag virus at a multiplicity of infection (MOI) of 2 and analyzed pM79 and pM91 expression by immunoblotting against the tags (Fig. 3B). pM91 was not detected at 8 h postinfection (hpi) but was expressed after 24 hpi in a pattern similar to that of pM79 (Fig. 3B). In addition, the expressions of both were significantly reduced when the viral DNA synthesis inhibitor PAA (200 μg/ml) was supplemented at 24 hpi (Fig. 3C), suggesting that viral DNA synthesis greatly enhanced the expression of both pM79 and pM91. These data indicate that pM91 expression follows early-late kinetics.
FIG 3.
Expression pattern and localization of pM91 and pM79 during infection. (A) Representative diagram of the modifications in the genome of the MCMV BAC pSM79HA91flag. Fragments encoding a 3×HA tag or 3×FLAG tag were inserted in frame at the 3′ end of the M79 or M91 coding sequence, respectively. (B) pM91 and pM79 expression patterns. MEF10.1 cells were infected with the SM79HA91flag virus at an MOI of 2. Cells were harvested at the indicated hours postinfection (hpi), lysed, and analyzed by immunoblotting. pM91 was detected with anti-FLAG mouse antibodies, and pM79 was detected with anti-HA antibodies; α-tubulin was included as a loading control. (C) pM91 and pM79 expression and viral DNA synthesis. MEF10.1 cells were infected with the SM79HA91flag virus at an MOI of 2 in the presence or absence of the viral DNA synthesis inhibitor PAA (200 μg/ml). At 24 hpi, cells were harvested and analyzed as described in the legend to panel A. (D) pM91 colocalized with pM79. MEF10.1 cells were infected with SMgfp or SM79HA91flag virus at an MOI of 2, fixed at 24 hpi, and subjected to immunostaining with the rabbit anti-FLAG antibody and mouse anti-HA antibody to detect pM91 (green) and pM79 (red), respectively. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize the nuclei (blue). Bars = 10 μm.
We next examined the intracellular localization of pM91 and pM79 during infection. MEF10.1 cells infected with the SM79HA91flag virus were fixed at 24 hpi and subjected to an immunofluorescence assay with rabbit anti-FLAG and mouse anti-HA antibodies. Results showed that pM91 localized in the nucleus and was strongly associated with pM79 (Fig. 3D), indicating that pM91 colocalized with pM79 in replication compartments.
Collectively, we conclude that pM91 has similar expression kinetics as pM79 and that it colocalizes with pM79 during infection.
pM91 is essential for MCMV replication.
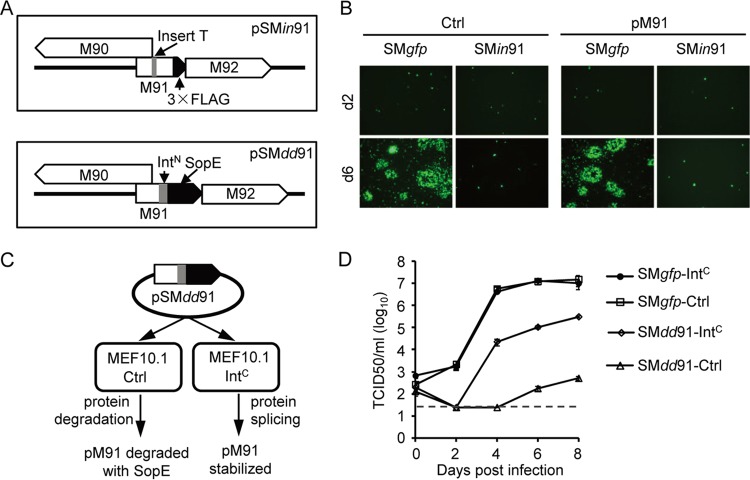
MCMV pM79, HCMV pUL79, and HCMV pUL91 were shown to be essential for viral late gene expression (16, 27, 29). As a pM79 partner, pM91 is also likely to be involved in this process. To understand the role of pM91 during infection, we engineered a pM91-deficient virus. Considering that the 5′-flanking region of the M91 coding sequence overlaps part of M90, we were unable to delete the entire open reading frame of pM91. Instead, we constructed a recombinant bacterial artificial chromosome (BAC), pSMin91, in which pM91 translation was disrupted by a premature stop codon by an insertional/frameshift mutation without interference with pM90 expression (Fig. 4A). We then transfected wild-type BAC pSMgfp or recombinant BAC pSMin91 into MEF10.1 cells. Both BACs were successfully transfected into MEF10.1 cells, with green fluorescent protein (GFP)-positive cells being present at 2 days posttransfection (Fig. 4B). However, only SMgfp spread to form plaques at 6 days posttransfection, whereas SMin91 produced only GFP-positive cells, indicating that this mutant virus had a significant replication deficiency (Fig. 4B, left). Unfortunately, the virus production of SMin91 was not rescued by pM91 expression in trans (Fig. 4B, right), and no mutant virus could be collected for further studies.
FIG 4.
M91 is essential for MCMV replication in fibroblasts. (A) Representative diagram of the pM91-deficient recombinant BACs pSMin91 and pSMdd91. pSMin91 has a single nucleotide insertion (T before nt 210) (indicated by the gray box) within the M91 coding sequence, resulting in a frameshift mutation. pSMdd91 contains the SopE coding sequence (indicated by the black box) and part of the gp41-1 split intein (IntN; indicated by the gray box) at the 3′ end of the M91 coding sequence. (B) Control (Ctrl) or pM91-expressing (pM91) MEF10.1 cells were transfected with wild-type BAC pSMgfp or mutant BAC pSMin91, and GFP signals were observed using fluorescence microscopy. (C) Working model of the pM91-deficient virus using the intein-mediated modulation of the protein stability system. Without IntC, SopE induces rapid degradation of the entire fusion protein, effectively depleting pM91 during infection. However, after intein splicing, SopE is released from pM91 and pM91 expression is rescued, supporting viral growth. (D) Control (Ctrl) or HA-mCherry-IntC-FLAG (short for IntC)-expressing MEF10.1 cells were infected with SMgfp or SMdd91 at an MOI of 0.01. At the indicated time points, the supernatants of infected cells were harvested and analyzed by the 50% tissue culture infective dose (TCID50) assay using MEF10.1-IntC cells. The detection limit of the TCID50 assay is indicated by the dashed line.
To overcome this problem, we created another recombinant BAC, pSMdd91, with an alternative approach using our previously reported intein-mediated modulation of protein stability (imPS) system (38). The intein domain is divided into N- and C-terminal fragments (IntN and IntC, respectively), each of which is fused to an external protein sequence (extein). IntN and IntC could associate and refold into an active intein, mediating protein trans-splicing and thus ligating the two exteins. In BAC pSMdd91, a fragment encoding a protein-destabilizing domain, SopE, and IntN were inserted in frame at the 3′ end of the M91 coding sequence, just upstream of the stop codon (Fig. 4A). Theoretically, in the cells with empty vectors (MEF10.1-Ctrl cells), pM91 should be degraded with SopE. On the contrary, in the presence of IntC, protein splicing should remove SopE from pM91 and stabilize the protein (Fig. 4C). As expected, we successfully harvested the mutant virus with HA-mCherry-IntC-FLAG-expressing MEF10.1 cells (MEF10.1-IntC cells).
To determine the replication capacity of the SMdd91 recombinant viruses, we measured multiple-step growth curves. MEF10.1-Ctrl cells or MEF10.1-IntC cells were infected with the SMgfp or SMdd91 virus at an MOI of 0.01, and the supernatants were collected at the various time points indicated below. The viral titer, determined by a 50% tissue culture infective dose (TCID50) assay, was measured using MEF10.1-IntC cells. As shown in Fig. 4D, the growth of the wild-type virus (SMgfp) in MEF10.1-IntC cells was similar to that in MEF10.1-Ctrl cells, indicating that HA-mCherry-IntC-FLAG expression does not affect SMgfp virus growth. In contrast, the recombinant virus SMdd91 almost failed to grow in the MEF10.1-Ctrl cells, as the levels of the viruses collected at 8 days postinfection (dpi) were almost undetectable (5 × 102 TCID50 units/ml). Nevertheless the levels of the viruses collected from SMdd91-infected MEF10.1-IntC cells reached 3 × 105 TCID50 units/ml at 8 dpi. Although the level of growth of the recombinant virus SMdd91 in complementary cells (MEF10.1-IntC) was not rescued to that of the wild-type virus SMgfp, the imPS system indeed successfully controlled the stability of pM91 during virus infection and provided a unique approach to study the function of this essential gene.
Taken together, our findings suggest that pM91 is a factor essential for MCMV replication.
pM91 is required for late viral gene expression but dispensable for viral DNA replication.
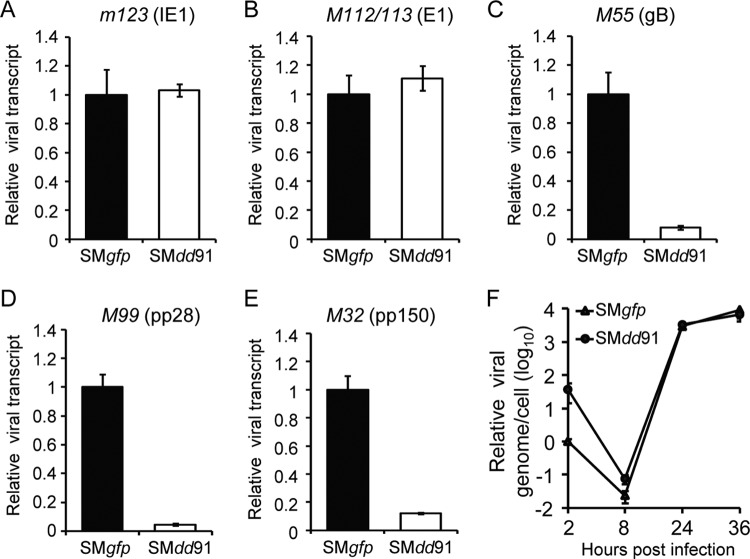
Using the pM91-deficient virus (SMdd91), we investigated the specific role of pM91 during infection. First, we examined the viral gene expression profile with reverse transcription (RT)-coupled quantitative PCR (qPCR) analysis. MEF10.1 cells were infected with pM91-deficient virus (SMdd91) or wild-type virus (SMgfp) at an MOI of 2. Total RNA was extracted to assess the RNA levels of the genes indicated below. At the early time point (8 hpi), depletion of pM91 had a minimal impact on immediate early gene m123 (IE1) or early gene M112/113 (E1) transcripts (Fig. 5A and B). However, at the late time spot (36 hpi), the levels of three important late gene transcripts, M55 (gB), M99 (pp28), and M32 (pp150), were 12.4-, 21.9-, and 8.3-fold lower, respectively, when pM91 was absent (Fig. 5C to E). The results suggest that pM91 is required for viral late gene transcription.
FIG 5.
pM91 is required for late gene expression but dispensable for viral lytic DNA replication. MEF10.1 cells were infected with wild-type virus (SMgfp) or pM91-deficient virus (SMdd91) at an MOI of 2. At 8 hpi (A, B) or 36 hpi (C, D, E), total RNA was extracted from the harvested cells and subjected to RT-qPCR. (A to E) The mRNA expression levels of the viral genes m123 (immediate early gene; IE1), M112/113 (early gene; E1), and M55 (gB), M99 (pp28), and M32 (pp150) (late genes) were normalized to the mouse Gapdh gene expression level, and the normalized amount of transcript during SMgfp infection was set to 1. (F) Depletion of pM91 had a minimal effect on viral DNA synthesis. MEF10.1 cells were infected as described in the legend to panel A, and intracellular DNA was isolated from infected cells at the indicated times. DNA levels were analyzed by qPCR, and the values were normalized to those for the actin gene. The quantity of DNA from SMgfp virus-infected cells at 2 hpi was set to 1.
As viral late gene transcription is activated by DNA replication, it is possible that pM91 inhibits DNA replication and further reduces late gene expression. To determine whether viral DNA replication was inhibited in the absence of pM91, we examined intracellular viral genomic copies in MEF10.1 cells infected with SMdd91 or SMgfp viruses at an MOI of 2. Total intracellular DNA was purified at the time points indicated below and subjected to qPCR analysis. The kinetics of viral genomic copies during SMdd91 infection were indistinguishable from those during SMgfp infection, suggesting that pM91 is dispensable for viral DNA replication (Fig. 5F).
The pM91-pM79 interaction is critical for virus production.
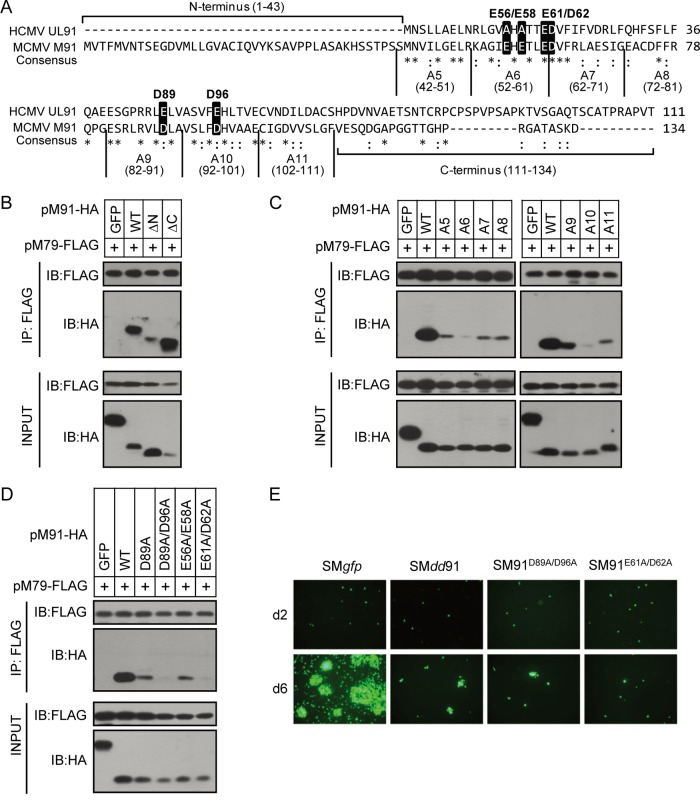
Next, we decided to map the pM79-interacting region within pM91. As mentioned above, pM91 and its homologs are conserved among beta- and gammaherpesviruses. Protein sequence alignment of pM91 (MCMV) and pUL91 (HCMV) revealed that the central regions were highly conserved, while the N- and C-terminal regions were less so (Fig. 6A). Amino acids (aa) 1 to 71 of pUL91 but not the C-terminal region was previously shown to be required for HCMV replication (27). Thus, truncation mutations were first generated to define which region of pM91 was responsible for the interaction. Neither N- nor C-terminal truncation had a significant effect on the interaction, suggesting that the central region was responsible (Fig. 6B). We then performed a 10-alanine screening toward the central region. Interestingly, all the pM91 alanine mutants (A5 to A11) greatly reduced the interaction, and the A6 and A10 mutants nearly abrogated binding (Fig. 6C). These results indicate that aa 52 to 61 and aa 92 to 101 of pM91 are critical for its interaction with pM79.
FIG 6.
Identification of the functional regions of pM91 for its interaction with pM79. (A) Protein sequence alignment of pM91 (MCMV) and the HCMV homolog pUL91. Identical and conserved amino acids are represented by asterisks and colons, respectively. The alanine scanning mutants used in panel C are also labeled. The amino acids in the black boxes indicate the positions mutated in panel D. (B, C, D) HEK293T cells were cotransfected with FLAG-tagged pM79 and either HA-tagged GFP (negative control), pM91 (wild type [WT]), or mutant pM91. Cell lysates were used for immunoprecipitation (IP) with anti-FLAG M2 beads, followed by immunoblotting (IB) with the indicated antibodies. pM91 truncated mutations included the N-terminus deletion (ΔN) and the C-terminus deletion (ΔC). A5 to A11 are pM91 alanine scanning mutants. D89A, D89A/D96A, E56A/E58A, and E61A/D62A indicate pM91 point mutants. (E) MEF10.1 cells were transfected with wild-type BAC pSMgfp or pSMdd91, pSM91D89A/D96A, and pSM91E61A/D62A mutant BACs, and the GFP signals were observed using fluorescence microscopy.
We also found that most residues in the central region of pM91 were charged. Interestingly, the predicted isoelectric point (pI) of pM91 is 4.81, suggesting that it has a strong negative charge under neutral pH. In contrast, pM79 (pI 9.37) has a strong positive charge, implying that the negatively charged residues within pM91 likely contribute to the interaction between these two proteins. Therefore, we mutated four conserved negatively charged residues (E61, D62, D89, and D96) within the pM91 central region. Strikingly, the D89A/D96A or E61A/D62A double mutation greatly reduced the interaction with pM79. On the contrary, mutation of D89A and double mutation of the less conserved negatively charged residues E56A/E58A had no significant effects on the interaction between pM91 and pM79 (Fig. 6D). This result suggests that the four residues E61, D62, D89, and D96 in pM91 are crucial for the pM91-pM79 interaction.
To dissect the role of the pM91-pM79 interaction during infection, we mutated these critical residues in the context of the viral genome to cause an interaction deficiency. After transfection of the BAC mutants (pSM91D89A/D96A or pSM91E61A/D62A) into MEF10.1 cells, phenotypes similar to those found after pM91-deficient BAC (pSMdd91) transfection were observed (Fig. 6E). The results indicate that the pM91-pM79 interaction is critical for viral production, likely through the regulation of viral late gene expression.
DISCUSSION
Late gene expression is a critical step to complete the life cycle of CMV lytic replication. Elucidating the regulation mechanism of this process is beneficial to explore novel antiviral strategies. In this study, we demonstrated that the six MCMV vTFs (pM49, pM79, pM87, pM91, pM92, and pM95) interacted directly or indirectly with each other to form a complex, in which pM91 had a strong direct interaction with pM79 (Fig. 1 and 2). We then revealed that pM91 is expressed with kinetics similar to those of pM79 and colocalizes with pM79 in the nuclear viral replication compartments during infection (Fig. 3). Depletion of pM91 inhibited late gene transcription and virus production but had only minor effects on viral DNA replication (Fig. 4 and 5). Finally, we mapped the essential residues within pM91 for pM79 binding, identifying four conserved residues, E61, D62, D89, and D96, which were essential for the interaction between these two proteins (Fig. 6). Mutation of these residues disrupted the pM91-pM79 interaction and impaired virus production (Fig. 6E). Taken together, the pM91-pM79 interaction is critical for the formation of the viral transcription complex and plays an essential role in the regulation of viral late gene expression.
Our study provides a model to explain the organization of the MCMV vTF complex (Fig. 2D). In brief, the six viral proteins form a complex by two sets of interactions: one with a weak interaction between pM92 and pM87 (Fig. 2A) and one with strong interactions: pM87-pM95, pM95-pM49, and pM91-pM79 (Fig. 2B). In addition, two of these subunits (pM95-pM49 and pM91-pM79) strongly interact with each other (Fig. 2C) and bind to pM87 through multiple interfaces. Recently, the EBV BcRF1 protein and KSHV ORF24 protein were reported to be TBP homologs (33, 34). As they are conserved in betaherpesviruses, pM87 probably also recognizes the TATT motif within late gene promoters and recruits other proteins to initiate viral late gene expression. It is interesting that although pM91 does not directly interact with pM87, pM95, or pM49 (Fig. 2B), it could coimmunoprecipitate with pM87, pM95, and pM49 with the presence of pM79 (Fig. 2C). One possible explanation is that the formation of the strong interactions (pM87-pM95, pM95-pM49, and pM91-pM79) likely changes the conformation of these proteins and generates new binding sites. These findings indicate the complexity of the vTF complex, and further structural analysis is needed to elucidate this detailed organization.
A previous study proved that KSHV ORF31-ORF34-ORF24 (pM92-pM95-pM87 in MCMV) forms a trimeric complex and ORF34 bridges the interaction between ORF31 and ORF24 (35). However, our results suggest that pM87 serves as the bridge to connect pM92 and pM95 (Fig. 2D). Another intriguing observation was that pM91 expression in trans could not rescue the replication of the pM91-deficient virus (Fig. 4B), while pUL91 expression could rescue the replication of the pUL91-null virus (27). These findings suggest that the conserved vTFs in beta- and gammaherpesviruses may possess different specificities and mechanisms.
Our study demonstrated that the pM91-pM79 interaction is required for formation of the vTF complex (Fig. 2C) and is essential for virus production (Fig. 6E). In addition, both pM91 (Fig. 5) and pM79 (16) are critical for viral late gene expression. It is likely that they have to interact with each other and promote the correct formation of the vTF complex during MCMV infection; otherwise, late gene transcription would be inhibited and progeny virus could not be produced. In the future, a pM91-pM79 interaction-deficient virus is needed to confirm this hypothesis.
In summary, our results shed insight into the organization of the viral late gene transcription complex during MCMV infection. Furthermore, we demonstrate the importance of pM91 and its interaction with pM79, which may provide novel strategies for the treatment of CMV-related diseases in the future.
MATERIALS AND METHODS
Plasmids and reagents.
MCMV M49, M79, M87, M91, M92, and M95 coding fragments were amplified from bacterial artificial chromosome (BAC) pSMgfp (16) and subcloned into pRetro-EBNA (39), modified to include N- or C-terminal tags (3×FLAG, 3×HA, or 3×Myc). Plasmid pEBNA-HA-mCherry-IntC-FLAG was generated by inserting the HA-mCherry-IntC-FLAG fragment (amplified from pLKO-dcMV-IntC-FLAG [38]) into pRetro-EBNA. Plasmid pYD-C163 is a pRetro-EBNA-based vector containing the coding sequence of HCMV protein pUL38 (40).
Plasmid pEPKan-S2 (41), which carries a kanamycin selection cassette bracketed by two I-SceI sites, was used for BAC recombination (42). To facilitate generation of the PCR product for BAC recombination, another three plasmids were constructed. The fragment KanS was amplified from pEPKan-S2 and inserted into the pEBNA vector with an N-terminal 3×HA tag or 3×FLAG tag, resulting in plasmids pEBNA-HA-KanS and pEBNA-FLAG-KanS. To create plasmid pEBNA-Myc-KanS-IntN-SopE, the fragment Myc-EcoRI-IntN-SopE was amplified from pLKO-dcMV-GFP-IntN-SopE (38) and inserted into pRetro-EBNA, and then the fragment KanS was inserted into the EcoRI site.
The primary antibodies used in this study included the following: anti-α-tubulin (66031-1-Ig; Proteintech), anti-FLAG (M20008; Abmart), anti-FLAG (F7425; Sigma-Aldrich), anti-HA (MMS-101P; Covance), anti-MCMV IE1 and E1 (generous gifts from Stipan Jonjic, University of Rijeka, Rijeka, Croatia), anti-MCMV gB (a generous gift from Anthony Scalzo, University of Western Australia), and anti-pUL38 (a generous gift from Thomas Shenk, Princeton University). Other chemicals used in this study included phosphonoacetic acid (PAA; 284270-10G; Sigma-Aldrich) and l-(+)-arabinose (Urchem).
Cells.
Mouse embryonic fibroblast 10.1 (MEF10.1) cells (43), Phoenix cells (39), and HEK293T cells were propagated in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, nonessential amino acids, and 1 mM sodium pyruvate. To create MEF10.1 cells expressing pM91 (MEF10.1-pM91), the empty vector (MEF10.1-Ctrl), or HA-mCherry-IntC-FLAG (MEF10.1-IntC), retrovirus stocks were made by transfecting the retroviral vector pEBNA-pM91, pRetro-EBNA, or pEBNA-HA-mCherry-IntC-FLAG, respectively, into Phoenix cells. Then, MEF10.1 cells were transduced three times with these retroviruses as described previously (44).
BAC mutagenesis and recombinant viruses.
MCMV BAC pSMgfp (a kind gift from Dong Yu) carries the full-length genome of the MCMV Smith strain (45) with a GFP expression cassette at the C terminus of the MCMV IE2 locus (16). To modify the MCMV genome, pSMgfp was introduced into Escherichia coli strain GS1783 (a kind gift from Gregory Smith) by electroporation. All recombinant MCMV BAC clones used in this study were constructed according to a BAC recombineering protocol described previously (42). To generate BAC pSM79HA91flag, we first created BAC pSM79HA. pSM79HA was constructed by inserting an oligonucleotide encoding 3×HA at the 3′ end of the M79 coding sequence within pSMgfp. Then, the oligonucleotide encoding 3×FLAG was inserted at the 3′ end of the M91 coding sequence in pSM79HA, resulting in recombinant BAC pSM79HA91flag. BAC pSMin91 carried a single nucleotide insertion (T inserted before nucleotide [nt] 210) within the M91 coding sequence of pSM79HA91flag, resulting in a frameshift mutation. BAC pSMdd91 contained the SopE coding sequence and part of the gp41-1 split intein (IntN) at the 3′ end of the M91 coding sequence of pSMgfp. BAC pSM91D89A/D96A and pSM91E61A/D62A carried a double mutation in the M91 coding sequence of pSM79HA91flag. The primers used for mutagenesis are listed in Table 2.
TABLE 2.
Primers used in BAC recombination
| Primer namea | Sequence (5′–3′)b |
|---|---|
| M91-FLAG-F | cgccgggcggaacgacgggccaccctcgcggagcgaccgcctcgaaggacGACTACAAAGACCATGACGGTG |
| M91-FLAG-R | cgatataggcttgcccgtccgcgggtctcggccgtgtgagaacatagtcaCTTGTCATCGTCATCCTTGTAATCG |
| M79-HA-F | acaagaaggaggaaggtgtgatcgccgacctgctgagcggagcgctcgaaTACCCTTATGACGTGCCCGA |
| M79-HA-R | gagatataccgtgtgtgggtcgtttttttttttattcataaaatcaatcaAGCATAATCTGGAACATCATATGG |
| M91-dd-F | cgccgggcggaacgacgggccaccctcgcggagcgaccgcctcgaaggacGAGCAAAAGCTGATCTCCGAGGAG |
| M91-dd-R | cgatataggcttgcccgtccgcgggtctcggccgtgtgagaacatagtcaGGCGCTGCCGCGGATGTCG |
| M91-inT-F | acgagaccctggaagacgtgtttcgtctcgccgagtccatTcggggaggcgtgcgacttctAGGATGACGACGATAAGTAGGG |
| M91-inT-R | ggattccccgggttgcctgaagaagtcgcacgcctccccgAatggactcggcgagacgaaaCAACCAATTAACCAATTCTGATTAG |
| M91-D89A/D96A-F | gtgcgacttcttcaggcaacccggggaatcccgactgcgggtcttgGCCctcgcggtgtccttatttGCCcacgttgcggcggagtgtatAGGATGACGACGATAAGTAGGG |
| M91-D89A/D96A-R | gaagcccagcgaaaccacgtctcctatacactccgccgcaacgtgGGCaaataaggacaccgcgagCAACCAATTAACCAATTCTGATTAG |
| M91-E61A/D62A-F | cggcgagctcaggaaggccggcatcgagcacgagaccctgGCCGCTgtgtttcgtctcgccgagtcAGGATGACGACGATAAGTAGGG |
| M91-E61A/D62A-R | agtcgcacgcctccccgatggactcggcgagacgaaacacAGCGGCcagggtctcgtgctCAACCAATTAACCAATTCTGATTAG |
F, forward; R, reverse.
Lowercase nucleotides indicate sequences homologous to parental BACs, underlined uppercase nucleotides indicate mutagenesis site, and uppercase nucleotides indicate sequences homologous to those of the plasmids used as a template.
The above-described BAC clones were then used to reconstitute the corresponding viruses. Two to 5 μg of MCMV BAC DNA was electroporated into normal MEF10.1 cells or MEF10.1-IntC cells (for reconstituted SMdd91) and plated on a 10-cm plate. The culture medium was changed after 24 h, and virus stocks were prepared by harvesting cell-free culture supernatant when 80% of the infected cells were lysed. Viruses were amplified by infecting cells at a multiplicity of infection (MOI) of 0.01 and collecting the cell-free supernatant from infected cultures. Virus titers were determined in duplicate by a 50% tissue culture infective dose (TCID50) assay in MEF10.1 cells or MEF10.1-IntC cells.
Viral growth analysis.
The BAC-transfected cells were observed with a Leica fluorescence microscope. Images were captured with a Leica DFC 450 digital camera. The growth kinetics of virus SMdd91 were analyzed as previously described (44). MEF10.1-Ctrl cells or MEF10.1-IntC cells were seeded in 12-well dishes. After 24 h, the cells were inoculated with SMdd91 virus for 1 h at an MOI of 0.01. The inoculum was removed, the cells were rinsed with fresh medium, and finally, 1 ml fresh medium was added to each well. Cell-free media from infected cultures were collected in duplicate at various times postinfection, and the virus titers in the media were determined by the TCID50 assay in MEF10.1-IntC cells.
Protein analysis.
Protein interactions were analyzed by coimmunoprecipitation assay as previously described (30). HEK293T cells were transfected with the plasmids indicated above and collected after 48 h. Collected cells were lysed in 2 ml lysis buffer (40 mM HEPES [pH 7.4], 1 mM EDTA, 300 mM NaCl, 0.5% NP-40) supplemented with 250 units of Benzonase nuclease (which digested the DNA and prevented DNA-mediated interactions; Millipore), phenylmethylsulfonyl fluoride, and protease inhibitors (PIC; Roche), incubated at 4°C for 1 h, and centrifuged at 13,200 × g at 4°C for 15 min. Fifty microliters of the supernatant was saved as the input control and boiled in sodium dodecyl sulfate (SDS)-containing sample buffer. The remainder was incubated with FLAG M2 (mouse monoclonal) antibody-conjugated magnetic beads (Sigma-Aldrich) at 4°C for 4 h. Then, the beads were washed four times with 2 ml lysis buffer. The immunoprecipitants were eluted by 150 ng/μl FLAG peptide (Sigma-Aldrich). The input and elution were analyzed by immunoblotting with the antibodies indicated above.
Protein accumulation was analyzed by immunoblotting as described previously (40). Briefly, cells were collected and lysed in SDS sample buffer. Proteins were resolved by SDS-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane, hybridized with primary antibodies, reacted with horseradish peroxidase-coupled secondary antibodies, and visualized by the use of Clarity Western enhanced chemiluminescence substrate (Bio-Rad).
The intracellular localization of the proteins of interest was analyzed by immunofluorescence assay as previously described (46). Cells were seeded onto coverslips and infected with SMgfp or SM79HA91flag at an MOI of 2. At 24 h postinfection (hpi), cells were washed, fixed, permeabilized, blocked, incubated with primary antibodies, and subsequently labeled with Alexa Fluor 488 (anti-mouse)- or Alexa Fluor 568 (anti-rabbit)-conjugated secondary antibodies (Invitrogen-Molecular Probes). Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Beyotime) and mounted on slides with Prolong Gold antifade reagent (Invitrogen-Molecular Probes). Confocal microscopic images were captured by an Olympus FV1200 confocal laser scanning microscope.
RNA and DNA analysis.
Relative mRNA levels were determined by reverse transcription (RT)-quantitative PCR (qPCR) as previously described (38). Total RNA was extracted with the TRIzol reagent (Invitrogen), and cDNA was reverse transcribed using a PrimeScript RT reagent kit (TaKaRa). cDNA was then quantified using SYBR Premix Ex Taq (TaKaRa) with primer pairs specific for viral genes or the mouse Gapdh (glyceraldehyde-3-phosphate dehydrogenase) gene (Table 3) (16). All reactions were performed in two biological and two technical replicates. The amounts of viral transcript were normalized to the amount of Gapdh.
TABLE 3.
Primers used in qPCR analysis
| Primera | Sequence (5′–3′) |
|---|---|
| MCMV m123 (IE1)-F | CAGGGTGGATCATGAAGCCT |
| MCMV m123 (IE1)-R | AGCGCATCGAAAGACAACG |
| MCMV M112/113 (E1)-F | GAATCCGAGGAGGAAGACGAT |
| MCMV M112/113 (E1)-R | GGTGAACGTTTGCTCGATCTC |
| MCMV M55 (gB)-F | GCGATGTCCGAGTGTGTCAAG |
| MCMV M55 (gB)-R | CGACCAGCGGTCTCGAATAAC |
| MCMV M99 (pp28)-F | GTCTGACGACGAAGACCAGG |
| MCMV M99 (pp28)-R | CTGTATCGGAGTGACCACGG |
| MCMV M32 (pp150)-F | GAGACGATGAGGATTGCGGT |
| MCMV M32 (pp150)-R | GGCCTCGTTGTCCACCATTA |
| Mouse Actin-F | GCTGTATTCCCCTCCATCGTG |
| Mouse Actin-R | CACGGTTGGCCTTAGGGTTCA |
| Mouse Gapdh-F | TGGAGAAACCTGCCAAGTATGA |
| Mouse Gapdh-R | CTGTTGAAGTCGCAGGAGACAA |
F, forward; R, reverse.
Intracellular DNA was measured by qPCR as previously described (16). MEF10.1 cells were infected with the SMgfp virus or SMdd91 virus at an MOI of 2 and collected at the times postinfection indicated above. Viral or cellular DNA was quantified by qPCR using SYBR Premix Ex Taq (TaKaRa) with a primer pair specific for MCMV m123 (IE1) or the mouse actin gene, respectively (Table 3) (16). The accumulation of viral DNA was normalized by dividing the number of m123 gene equivalents by the number of actin gene equivalents. The normalized amount of viral DNA in SMgfp-infected cells at 2 hpi was set at 1.
ACKNOWLEDGMENTS
We thank all the members of the Herpesvirus and Molecular Virology Research Unit for helpful discussions and invaluable advice; Dong Yu (GSK Vaccines, USA), Martin Messerle (Hanover Medical School, Hanover, Germany), Wolfram Brune (Heinrich Pette Institute, Leibniz Institute for Experimental Virology, Hamburg, Germany), and Ulrich Koszinowski (Max von Pettenkofer Institute, Ludwig Maximilian University, Munich, Germany) for the MCMV BAC clone pSMgfp; Anthony Scalzo (University of Western Australia) for the gB antibody; Stipan Jonjic (University of Rijeka, Rijeka, Croatia) for the IE1 and E1 antibodies; Thomas Shenk (Princeton University) for the pUL38 antibody; Gregory Smith (Northwestern University Feinberg School of Medicine) for E. coli GS1783; and the members of the Ke Lan (Wuhan University, China) lab for technical support with BAC recombination.
This work was supported by the National Natural Science Foundation of China (grants 81371826 and 81572002 to Z.Q., grants 31300148 and 31570169 to B.X.), the Ministry of Science and Technology of China (2016YFA0502101), and the Chinese Academy of Sciences (CAS) 100 Talents program to Z.Q. B.X. was supported by the Youth Innovation Promotion Association, CAS.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Davison AJ. 2007. Overview of classification. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 2.Crough T, Khanna R. 2009. Immunobiology of human cytomegalovirus: from bench to bedside. Clin Microbiol Rev 22:76–98. doi: 10.1128/CMR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mocarski ES, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2701–2772. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol II Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4.Nogalski MT, Collins-McMillen D, Yurochko AD. 2014. Overview of human cytomegalovirus pathogenesis. Methods Mol Biol 1119:15–28. doi: 10.1007/978-1-62703-788-4_2. [DOI] [PubMed] [Google Scholar]
- 5.Anderholm KM, Bierle CJ, Schleiss MR. 2016. Cytomegalovirus vaccines: current status and future prospects. Drugs 76:1625–1645. doi: 10.1007/s40265-016-0653-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biron KK. 2006. Antiviral drugs for cytomegalovirus diseases. Antiviral Res 71:154–163. doi: 10.1016/j.antiviral.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed A. 2011. Antiviral treatment of cytomegalovirus infection. Infect Disord Drug Targets 11:475–503. doi: 10.2174/187152611797636640. [DOI] [PubMed] [Google Scholar]
- 8.Mercorelli B, Sinigalia E, Loregian A, Palu G. 2008. Human cytomegalovirus DNA replication: antiviral targets and drugs. Rev Med Virol 18:177–210. doi: 10.1002/rmv.558. [DOI] [PubMed] [Google Scholar]
- 9.Krmpotic A, Bubic I, Polic B, Lucin P, Jonjic S. 2003. Pathogenesis of murine cytomegalovirus infection. Microbes Infect 5:1263–1277. doi: 10.1016/j.micinf.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Rawlinson WD, Farrell HE, Barrell BG. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol 70:8833–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeMarchi JM, Schmidt CA, Kaplan AS. 1980. Patterns of transcription of human cytomegalovirus in permissively infected cells. J Virol 35:277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keil GM, Ebeling-Keil A, Koszinowski UH. 1984. Temporal regulation of murine cytomegalovirus transcription and mapping of viral RNA synthesized at immediate early times after infection. J Virol 50:784–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stinski MF, Meier JL. 2007. Immediate-early viral gene regulation and function. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 14.White EA, Spector DH. 2007. Early viral gene expression and function. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 15.Huang ES. 1975. Human cytomegalovirus. IV. Specific inhibition of virus-induced DNA polymerase activity and viral DNA replication by phosphonoacetic acid. J Virol 16:1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapa TJ, Johnson LS, Affolter C, Valentine MC, Fehr AR, Yokoyama WM, Yu D. 2013. Murine cytomegalovirus protein pM79 is a key regulator for viral late transcription. J Virol 87:9135–9147. doi: 10.1128/JVI.00688-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anders DG, Kerry JA, Pari GS. 2007. DNA synthesis and late viral gene expression. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 18.Gong D, Wu NC, Xie Y, Feng J, Tong L, Brulois KF, Luan H, Du Y, Jung JU, Wang CY, Kang MK, Park NH, Sun R, Wu TT. 2014. Kaposi's sarcoma-associated herpesvirus ORF18 and ORF30 are essential for late gene expression during lytic replication. J Virol 88:11369–11382. doi: 10.1128/JVI.00793-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis ZH, Hesser CR, Park J, Glaunsinger BA. 2015. Interaction between ORF24 and ORF34 in the Kaposi's sarcoma-associated herpesvirus late gene transcription factor complex is essential for viral late gene expression. J Virol 90:599–604. doi: 10.1128/JVI.02157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aubry V, Mure F, Mariame B, Deschamps T, Wyrwicz LS, Manet E, Gruffat H. 2014. Epstein-Barr virus late gene transcription depends on the assembly of a virus-specific preinitiation complex. J Virol 88:12825–12838. doi: 10.1128/JVI.02139-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Djavadian R, Chiu YF, Johannsen E. 2016. An Epstein-Barr virus-encoded protein complex requires an origin of lytic replication in cis to mediate late gene transcription. PLoS Pathog 12:e1005718. doi: 10.1371/journal.ppat.1005718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu TT, Park T, Kim H, Tran T, Tong L, Martinez-Guzman D, Reyes N, Deng H, Sun R. 2009. ORF30 and ORF34 are essential for expression of late genes in murine gammaherpesvirus 68. J Virol 83:2265–2273. doi: 10.1128/JVI.01785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arumugaswami V, Wu TT, Martinez-Guzman D, Jia Q, Deng H, Reyes N, Sun R. 2006. ORF18 is a transfactor that is essential for late gene transcription of a gammaherpesvirus. J Virol 80:9730–9740. doi: 10.1128/JVI.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia Q, Wu TT, Liao HI, Chernishof V, Sun R. 2004. Murine gammaherpesvirus 68 open reading frame 31 is required for viral replication. J Virol 78:6610–6620. doi: 10.1128/JVI.78.12.6610-6620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davison AJ, Bhella D. 2007. Comparative genome and virion structure. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 26.Isomura H, Stinski MF, Murata T, Yamashita Y, Kanda T, Toyokuni S, Tsurumi T. 2011. The human cytomegalovirus gene products essential for late viral gene expression assemble into prereplication complexes before viral DNA replication. J Virol 85:6629–6644. doi: 10.1128/JVI.00384-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Omoto S, Mocarski ES. 2013. Cytomegalovirus UL91 is essential for transcription of viral true late (gamma2) genes. J Virol 87:8651–8664. doi: 10.1128/JVI.01052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omoto S, Mocarski ES. 2014. Transcription of true late (gamma2) cytomegalovirus genes requires UL92 function that is conserved among beta- and gammaherpesviruses. J Virol 88:120–130. doi: 10.1128/JVI.02983-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perng YC, Qian Z, Fehr AR, Xuan B, Yu D. 2011. The human cytomegalovirus gene UL79 is required for the accumulation of late viral transcripts. J Virol 85:4841–4852. doi: 10.1128/JVI.02344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perng YC, Campbell JA, Lenschow DJ, Yu D. 2014. Human cytomegalovirus pUL79 is an elongation factor of RNA polymerase II for viral gene transcription. PLoS Pathog 10:e1004350. doi: 10.1371/journal.ppat.1004350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapa TJ, Perng YC, French AR, Yu D. 2014. Murine cytomegalovirus protein pM92 is a conserved regulator of viral late gene expression. J Virol 88:131–142. doi: 10.1128/JVI.02684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyrwicz LS, Rychlewski L. 2007. Identification of herpes TATT-binding protein. Antiviral Res 75:167–172. doi: 10.1016/j.antiviral.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Davis ZH, Verschueren E, Jang GM, Kleffman K, Johnson JR, Park J, Von Dollen J, Maher MC, Johnson T, Newton W, Jager S, Shales M, Horner J, Hernandez RD, Krogan NJ, Glaunsinger BA. 2015. Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol Cell 57:349–360. doi: 10.1016/j.molcel.2014.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gruffat H, Kadjouf F, Mariame B, Manet E. 2012. The Epstein-Barr virus BcRF1 gene product is a TBP-like protein with an essential role in late gene expression. J Virol 86:6023–6032. doi: 10.1128/JVI.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brulois K, Wong LY, Lee HR, Sivadas P, Ensser A, Feng P, Gao SJ, Toth Z, Jung JU. 2015. Association of Kaposi's sarcoma-associated herpesvirus ORF31 with ORF34 and ORF24 is critical for late gene expression. J Virol 89:6148–6154. doi: 10.1128/JVI.00272-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishimura M, Watanabe T, Yagi S, Yamanaka T, Fujimuro M. 2017. Kaposi's sarcoma-associated herpesvirus ORF34 is essential for late gene expression and virus production. Sci Rep 7:329. doi: 10.1038/s41598-017-00401-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chapa TJ, Yu D, French AR. 2017. Proteomic and phylogenetic coevolution analyses of pM79 and pM92 identify interactions with RNA polymerase II and delineate the murine cytomegalovirus late transcription complex. J Gen Virol 98:242–250. doi: 10.1099/jgv.0.000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan D, Xuan B, Sun Y, Huang S, Xie M, Bai Y, Xu W, Qian Z. 2016. An intein-mediated modulation of protein stability system and its application to study human cytomegalovirus essential gene function. Sci Rep 6:26167. doi: 10.1038/srep26167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinsella TM, Nolan GP. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum Gene Ther 7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 40.Terhune S, Torigoi E, Moorman N, Silva M, Qian Z, Shenk T, Yu D. 2007. Human cytomegalovirus UL38 protein blocks apoptosis. J Virol 81:3109–3123. doi: 10.1128/JVI.02124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 42.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless Red recombination system. Methods Mol Biol 634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 43.Harvey DM, Levine AJ. 1991. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev 5:2375–2385. doi: 10.1101/gad.5.12b.2375. [DOI] [PubMed] [Google Scholar]
- 44.Qian Z, Xuan B, Chapa TJ, Gualberto N, Yu D. 2012. Murine cytomegalovirus targets transcription factor ATF4 to exploit the unfolded-protein response. J Virol 86:6712–6723. doi: 10.1128/JVI.00200-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner M, Jonjic S, Koszinowski UH, Messerle M. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol 73:7056–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie M, Xuan B, Shan J, Pan D, Sun Y, Shan Z, Zhang J, Yu D, Li B, Qian Z. 2015. Human cytomegalovirus exploits interferon-induced transmembrane proteins to facilitate morphogenesis of the virion assembly compartment. J Virol 89:3049–3061. doi: 10.1128/JVI.03416-14. [DOI] [PMC free article] [PubMed] [Google Scholar]