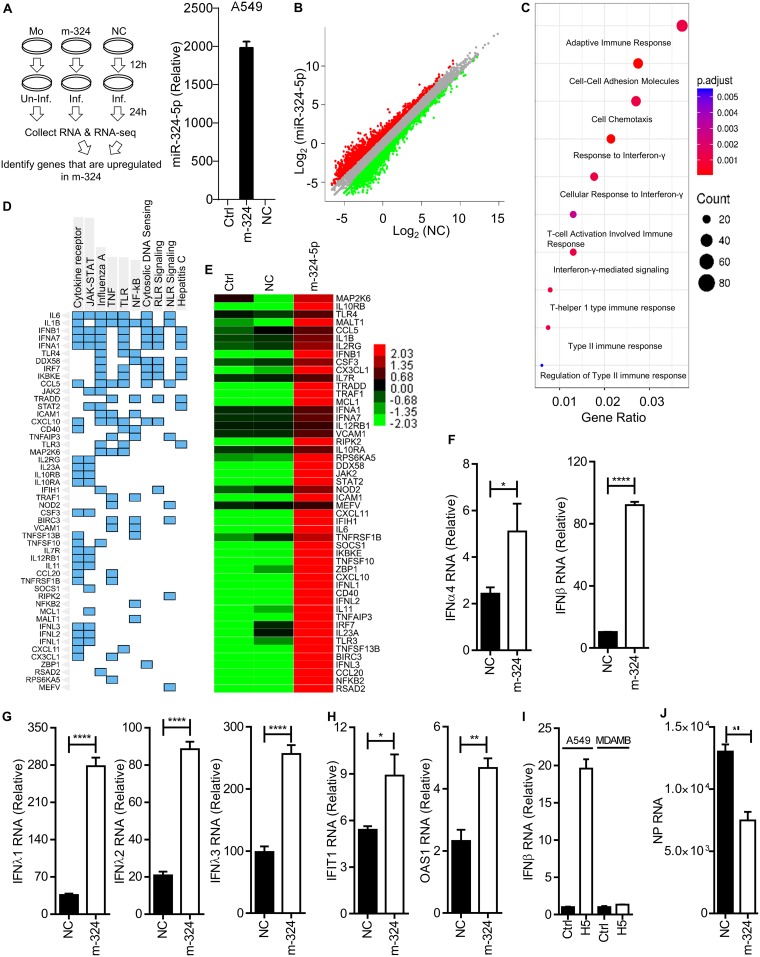
FIG 4.
miR-324-5p enhances ISGs and type I and type III interferons. (A) Schematic outline of transfection, infection (H5N1), RNA isolation, sequencing, and RNA-Seq analysis. Mo, mock; Inf., infected; Un-inf., uninfected. (B) Global expression analysis during m-324 overexpression in H5N1-infected A549 cells using R package. NC is plotted versus m-324 in H5N1-infected samples. Genes significantly changed (>2-fold) are colored in red and green for upregulated and downregulated, respectively. (C) Gene ontologies represented as dot plots. The circles are colored based on the significance (P value) of enrichment, while the sizes represent the number of genes that are enriched for that particular category. (D) Clustergram representing enriched KEGG pathways analysis as columns and input genes as rows; cells in the matrix (blue color) indicate if a gene is associated with the pathway. (E) Heat map analysis of upregulated genes associated with innate immune responses. (F to H) A549 cells were transfected with either m-324 or NC, followed by infection with H5N1 at an MOI of 5, and subjected to qRT-PCR analysis after 24 h to determine the relative expression of IFN-α4 and IFN-β (F), IFN-λ1, IFN-λ2, and IFN-λ3 (G), and IFIT1 and OAS1 (H) RNAs. (I) Equal numbers of A549 and MDAMB-231 cells were either left uninfected (Ctrl) or infected with H5N1 at an MOI of 1 for 24 h. IFN-β RNA levels were quantified by qRT-PCR. (J) MDAMB-231 cells were transfected with 100 nM m-324 or NC, followed by infection with H5N1 at an MOI of 1 for 24 h. Relative levels of NP RNA were quantified by qRT-PCR. Data are means ± SEMs of triplicate samples of a single experiment and are representative of results from three independent experiments (F to J). ****, P < 0.0001, ***, P < 0.001, **, P < 0.01, and *, P < 0.05, by two-tailed unpaired t test.