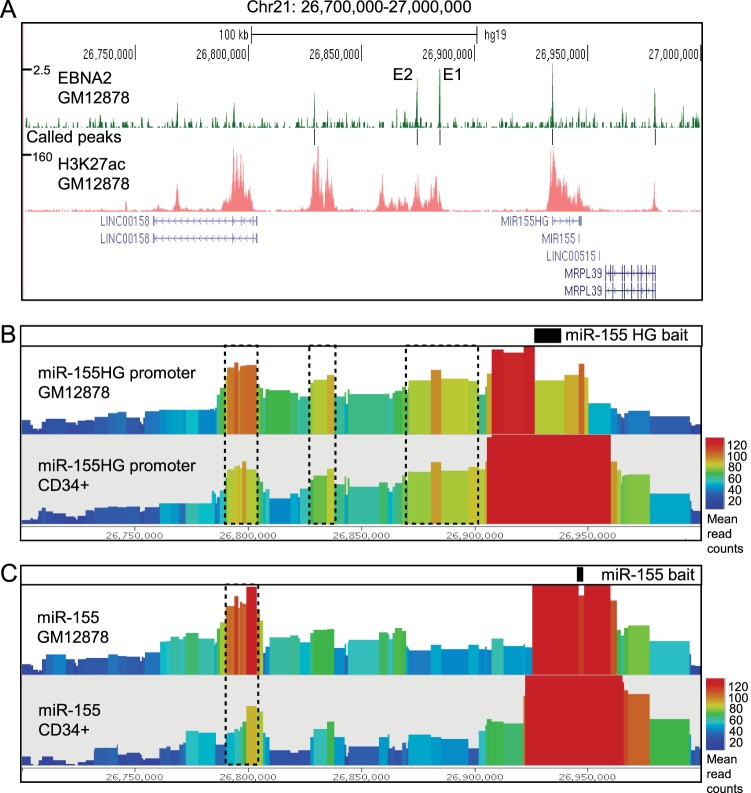
FIG 1.
EBNA2 binding and chromosome interactions at the miR-155HG locus on chromosome 21. (A) EBNA2 ChIP sequencing in the EBV-infected LCL GM12878 (23) showing the number of sequencing reads from EBNA2-enriched DNA plotted per million input-subtracted total reads and aligned with the human genome. The position of called peaks (model-based analysis of ChIP-seq [MACS], P < 10−7) is indicated (black lines). The positions of the two main EBNA2-bound putative enhancer regions are indicated (E1 and E2). H3K27ac signals in GM12878 ChIP-seq data available from ENCODE are also shown. (B) Capture Hi-C interaction data obtained using a HindIII fragment encompassing the miR-155HG promoter as bait. Interacting fragments were captured from Hi-C libraries generated from GM12878 or CD34+ progenitor cells (33). Data show the normalized accumulation of raw read counts of the interaction ditags anchored on the bait regions. The geometric mean shows the average number of reads for each fragment and its immediate neighboring fragments colored according to a rainbow scale. The main interacting regions are shown in boxes with dashed lines. (C) Capture Hi-C interaction data obtained using a HindIII fragment encompassing the miR-155 genomic locus as bait. The main interacting region is shown in the box with dashed lines.