Nitrification is a critical process for preventing ammonia toxicity in engineered biofilter environments. This work describes the cultivation and complete genome sequence of a novel AOA representative enriched from a freshwater aquarium biofilter. In addition, despite the common belief in the aquarium industry that AOB mediate ammonia oxidation, the present study suggests an in situ role for “Ca. Nitrosotenuis aquarius”-like AOA in freshwater aquarium biofilters.
KEYWORDS: Thaumarchaeota, ammonia-oxidizing archaea, nitrification, biological filtration, aquarium
ABSTRACT
Ammonia is a metabolic waste product excreted by aquatic organisms that causes toxicity when it accumulates. Aquaria and aquaculture systems therefore use biological filters that promote the growth of nitrifiers to convert ammonia to nitrate. Ammonia-oxidizing bacteria (AOB) have been isolated from aquarium biofilters and are available as commercial supplements, but recent evidence suggests that ammonia-oxidizing archaea (AOA) are abundant in aquarium biofilters. In this study, we report the cultivation and closed genome sequence of the novel AOA representative “Candidatus Nitrosotenuis aquarius,” which was enriched from a freshwater aquarium biofilter. “Ca. Nitrosotenuis aquarius” oxidizes ammonia stoichiometrically to nitrite with a concomitant increase in thaumarchaeotal cells and a generation time of 34.9 h. “Ca. Nitrosotenuis aquarius” has an optimal growth temperature of 33°C, tolerates up to 3 mM NH4Cl, and grows optimally at 0.05% salinity. Transmission electron microscopy revealed that “Ca. Nitrosotenuis aquarius” cells are rod shaped, with a diameter of ∼0.4 μm and length ranging from 0.6 to 3.6 μm. In addition, these cells possess surface layers (S-layers) and multiple proteinaceous appendages. Phylogenetically, “Ca. Nitrosotenuis aquarius” belongs to the group I.1a Thaumarchaeota, clustering with environmental sequences from freshwater aquarium biofilters, aquaculture systems, and wastewater treatment plants. The complete 1.70-Mbp genome contains genes involved in ammonia oxidation, bicarbonate assimilation, flagellum synthesis, chemotaxis, S-layer production, defense, and protein glycosylation. Incubations with differential inhibitors indicate that “Ca. Nitrosotenuis aquarius”-like AOA contribute to ammonia oxidation within the aquarium biofilter from which it originated.
IMPORTANCE Nitrification is a critical process for preventing ammonia toxicity in engineered biofilter environments. This work describes the cultivation and complete genome sequence of a novel AOA representative enriched from a freshwater aquarium biofilter. In addition, despite the common belief in the aquarium industry that AOB mediate ammonia oxidation, the present study suggests an in situ role for “Ca. Nitrosotenuis aquarius”-like AOA in freshwater aquarium biofilters.
INTRODUCTION
Ammonia is a metabolic waste product excreted by fish and other aquatic organisms. Ammonia toxicity is of particular concern for relatively closed ecosystems, such as aquaculture operations and home aquaria, where ammonia can accumulate to stressful or lethal concentrations in the absence of sufficient nitrification. Biological filters help maintain low ammonia concentrations in these systems by promoting the growth of nitrifying populations, with high surface areas of filter support material (e.g., sponge, ceramic, and polymer) and rapid flows of aerated water.
Until recently, ammonia-oxidizing bacteria (AOB) were assumed to be solely responsible for ammonia oxidation in aquaria. Commercial filter supplements are commonly used to promote nitrification, and all supplements examined to date contain AOB (1, 2). In addition, AOB of the Nitrosomonas europaea, Nitrosospira tenuis, and Nitrosomonas marina lineages have been enriched from aquaculture environments (3). However, a study of 38 freshwater aquaria using oligonucleotide probes failed to detect AOB in all but two samples, suggesting that yet-unknown organisms must be responsible for ammonia oxidation in situ (4). Following the initial cultivation of “Candidatus Nitrosopumilus maritimus” as the first reported member of the ammonia-oxidizing archaea (AOA) (5), Urakawa and colleagues (6) detected amoA genes from both AOB and AOA in marine aquarium biofilters from a public aquarium in Japan and suggested that the diversity of AOA and AOB was decreased in low-temperature marine aquaria. Recent studies have demonstrated that AOA are numerically dominant over AOB in the majority of freshwater aquarium biofilters, implying an important role for AOA in aquarium nitrification (1, 2). AOA have also been detected in several aquaculture systems, including subarctic, temperate, and subtropical closed marine fish aquaculture systems (7), shrimp ponds in Thailand (8), and all compartments (including plastic bioballs, corrugated blocks, and crushed oyster shells) of a nitrifying trickling filter in a zero-discharge shrimp recirculating aquaculture system (9).
The first reported AOA representative, “Ca. Nitrosopumilus maritimus,” was isolated from a marine aquarium (5). As a model organism for marine AOA, “Ca. Nitrosopumilus maritimus” has provided insights into AOA ecology and physiology, such as demonstrations of high substrate affinities for ammonia (10), sensitivity to light (11), cell division by the Cdv system (12–14), and usage of hydroxylamine (15) and nitric oxide (16) as intermediates in ammonia oxidation. Despite the importance of freshwater ecosystems to human and environmental health, few freshwater AOA representatives exist. “Candidatus Nitrosoarchaeum limnia” is a low-salinity AOA representative originating from an estuary (17, 18). “Candidatus Nitrosotenuis uzonensis” originates from a freshwater hot spring and is moderately thermophilic (19). Within the same lineage, “Candidatus Nitrosotenuis chungbukensis” was enriched from a deep-soil horizon (20) and “Candidatus Nitrosotenuis cloacae” was enriched from a low-salinity wastewater treatment plant (WWTP) (20).
In this study, we report the cultivation and complete genome sequence of an AOA representative originating from a freshwater aquarium biofilter that was previously shown to have a high abundance of AOA (1). In addition, we demonstrate with differential inhibitors that AOA in aquarium biofilters contribute to nitrification in situ.
RESULTS AND DISCUSSION
“Candidatus Nitrosotenuis aquarius” enrichment culture.
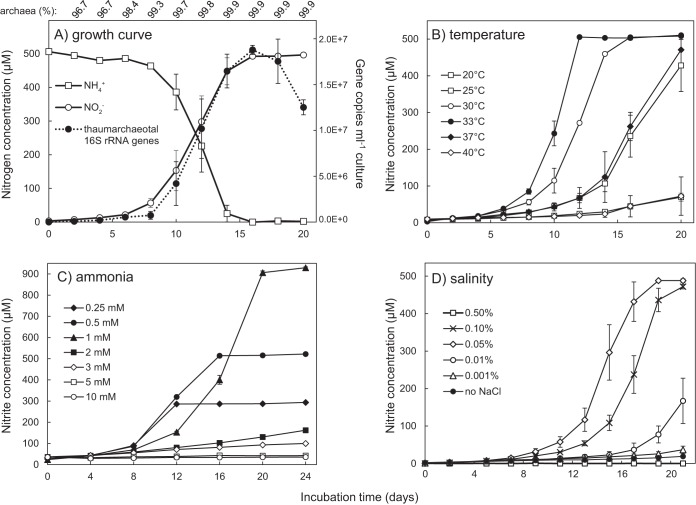
AOA may be important contributors to nitrogen and carbon cycling in freshwater environments, but few laboratory cultures of freshwater AOA exist. Here, we describe the highly enriched culture and complete genome sequence of “Ca. Nitrosotenuis aquarius” (a.quar.i.us. N.L. masc. adj. aquarius of or pertaining to water), an ammonia-oxidizing archaeon from a freshwater aquarium biofilter. “Ca. Nitrosotenuis aquarius” oxidizes ammonia to nitrite at near-stoichiometric values (Fig. 1A). Following a 1% subculture, an increase in thaumarchaeotal cell numbers (as estimated by 16S rRNA gene copy numbers) was observed concomitantly with nitrite production, providing evidence that growth is based on ammonia oxidation for energy conservation. The generation time of “Ca. Nitrosotenuis aquarius” at 30°C was 34.9 h, which is similar to that of “Ca. Nitrosopumilus maritimus” (33 h) (5), “Candidatus Nitrososphaera yellowstonii” (30 h) (21), and “Candidatus Nitrososphaera viennensis” (28 h) (22) and shorter than the generation times of “Ca. Nitrosotenuis cloacae” (70 h) (20), “Ca. Nitrosoarchaeum limnia” (82 h) (17), and “Candidatus Nitrosotalea devanaterra” (53 h) (23). Based on 16S rRNA gene abundances, the proportion of “Ca. Nitrosotenuis aquarius” in the enrichment culture ranged from 96.7 to 99.9% (Fig. 1A).
FIG 1.
Growth and ammonia-oxidizing activity of “Ca. Nitrosotenuis aquarius.” (A) Ammonia depletion and nitrite production in association with thaumarchaeotal cell numbers. Archaeal proportions (shown above the graph) correspond to time points below, and values are based on measured thaumarchaeotal and bacterial 16S rRNA gene copy numbers in genomic DNA extracts. Thaumarchaeotal 16S rRNA gene copy numbers represent an approximate number of “Ca. Nitrosotenuis aquarius” cells, assuming one gene copy per genome and one chromosome per cell. Subsequent panels show ammonia-oxidizing activity of “Ca. Nitrosotenuis aquarius” under various incubation temperatures (B), initial ammonia concentrations (C), and sodium chloride concentrations (D). Error bars represent the standard errors of biological triplicates for all panels; error bars not seen are contained within the symbols.
“Ca. Nitrosotenuis aquarius” actively produced nitrite at temperatures ranging from 20 to 40°C, with an optimal growth temperature of 33°C (Fig. 1B). None of the tested temperatures completely inhibited nitrite production although it was substantially slower at the lower (20°C) and upper (40°C) temperatures tested. Most group I.1a Thaumarchaeota are mesophiles with optimal growth temperatures ranging from 24 to 29°C (5, 17, 18, 23–25), but “Ca. Nitrosotenuis uzonensis” is moderately thermophilic, with an optimal growth temperature of 46°C (19). A slightly higher temperature range for “Ca. Nitrosotenuis aquarius” is consistent with its origin from a tropical fish tank (∼26 to 28°C), which is warmer than the typical marine and estuarine environments where many other group I.1a Thaumarchaeota originate.
“Ca. Nitrosotenuis aquarius” activity was highest when the archaeon was incubated with 0.25 to 1 mM NH4Cl (Fig. 1C), with partial inhibition of ammonia-oxidizing activity observed at 2 to 3 mM NH4Cl and complete inhibition at 5 mM NH4Cl. Similarly, the reported growth rate of “Ca. Nitrosotenuis cloacae” decreased at 3 mM, and growth was completely inhibited at 5 mM NH4Cl (24). When incubated with 0.5 mM urea as a sole energy source, “Ca. Nitrosotenuis aquarius” produced nitrite more slowly than cultures containing 0.5 mM ammonia (see Fig. S1A in the supplemental material). Accumulation of ammonia was not observed under urea-only conditions or in sterile controls (Fig. S1B). Given that urea supplementation did not result in equal rates of nitrite production, ammonia appears to be the preferred energy source of “Ca. Nitrosotenuis aquarius.” In an aquarium environment, some urea would be present from fish excretion (26, 27) and bacterial decomposition (28, 29), but the majority of nitrogenous waste excreted by fish would be in the form of ammonia.
“Ca. Nitrosotenuis aquarius” grew optimally at a salt concentration of 0.05% NaCl and tolerated up to 0.1% NaCl (Fig. 1D). “Ca. Nitrosotenuis uzonensis” also grows optimally at 0.05% NaCl (19), whereas “Ca. Nitrosotenuis cloacae” grows optimally at 0.03% salinity (24). In contrast, members of the “Candidatus Nitrosoarchaeum” lineage tolerate higher salt concentrations, with “Candidatus Nitrosoarchaeum koreensis,” for example, tolerating salinity up to 0.4% (25). “Ca. Nitrosoarchaeum limnia” grows optimally at 0.88% salinity and tolerates up to 2.6% salinity (18), which is consistent with its presence in higher-salinity environments, such as salt marshes. For comparison, the marine AOA representative “Ca. Nitrosopumilus maritimus” tolerates salinities of >3.5% (5).
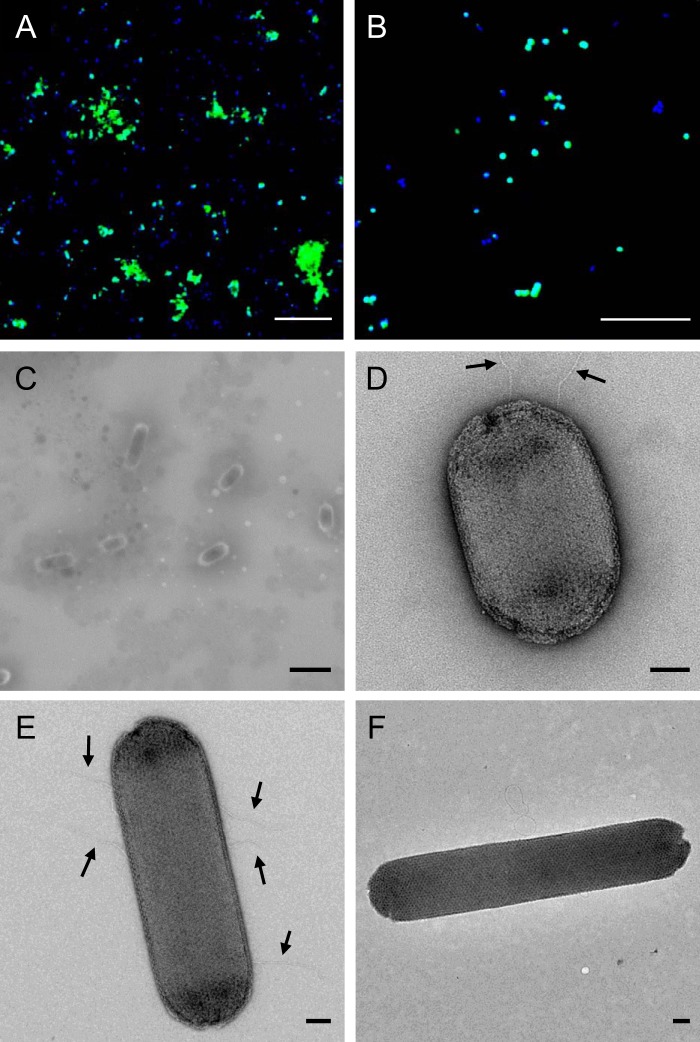
“Ca. Nitrosotenuis aquarius” cells detected by catalyzed reporter deposition-fluorescent in situ hybridization (CARD-FISH) were predominantly found in a clumped biomass (Fig. 2A). Although individual cells were small (∼0.5 μm in diameter) with a coccoid morphology (Fig. 2B), cells imaged by scanning electron microscopy (SEM) were slender rods with a diameter of ∼0.3 to 0.4 μm and length ranging from ∼0.5 to 1.5 μm (Fig. 2C). It is likely that apparently coccoid cells visualized by CARD-FISH were actually short rods (i.e., coccobacilli), given the comparatively low resolution of fluorescence microscopy. “Ca. Nitrosotenuis aquarius” cells imaged with transmission electron microscopy (TEM) displayed crystalline surface layers (S-layers) (Fig. 2D to F), and the majority of cells imaged possessed appendages (Fig. 2D and E), presumably flagella. Cells typically had five or fewer appendages, which ranged from subpolar localization (Fig. 2D) to localization around the cell periphery (Fig. 2E). These appendages were relatively short, but it is possible that they were damaged during processing. In TEM images, “Ca. Nitrosotenuis aquarius” cells were consistently 0.4 μm in diameter (Fig. 2D to F) and displayed a broad range of lengths, from very short rods or coccobacilli (0.6 μm in length) (Fig. 2D) up to 6-fold-longer rods (3.6 μm in length) (Fig. 2F).
FIG 2.
“Ca. Nitrosotenuis aquarius” micrographs. (A and B) CARD-FISH micrographs of enrichment cultures using the probe Arch915 (FITC tyramides; green), counterstained with DAPI (blue). Scale bar, 10 μm. (C) Scanning electron micrograph of unstained “Ca. Nitrosotenuis aquarius” cells. Scale bar, 1 μm. (D to F) Transmission electron micrographs of negatively stained “Ca. Nitrosotenuis aquarius” cells (2% uranyl acetate). Scale bar, 100 nm. Arrows in panel E indicate cellular appendages.
Rod and coccobacillus morphologies and small cell sizes have been reported for most other group I.1a Thaumarchaeota (5, 23, 25, 30). “Ca. Nitrosotenuis aquarius” shares several morphological similarities with “Ca. Nitrosotenuis uzonensis” (19) and “Ca. Nitrosoarchaeum limnia” (18). These cells demonstrate rod morphologies, with thin appendages presumed to be flagella and crystalline S-layers. Although several relatively short putative flagella were observed in “Ca. Nitrosotenuis aquarius,” only a single long flagellum was observed in “Ca. Nitrosoarchaeum limnia,” and one or two flagella per cell were found in “Ca. Nitrosotenuis uzonensis.” Despite similar morphologies, “Ca. Nitrosotenuis aquarius” cells were larger than those of other group I.1a Thaumarchaeota. For example, “Ca. Nitrosoarchaeum limnia” cells are 0.19 to 0.27 μm by 0.55 to 1.00 μm, and “Ca. Nitrosotenuis uzonensis” cells are 0.2 to 0.3 μm by 0.4 to 1.7. At up to 3.6 μm in length, “Ca. Nitrosotenuis aquarius” cells can grow to be up to 2-fold longer than “Ca. Nitrosotenuis uzonensis” cells and are the longest reported AOA cells to date. Interestingly, despite being closely related to “Ca. Nitrosotenuis aquarius,” “Ca. Nitrosotenuis cloacae” exhibits a strikingly different morphology; cells are coccoid, with a diameter of 1.1 ± 0.1 μm, and apparently lack any cellular appendages or S-layers (24). S-layers can have a variety of functions, including acting as molecular sieves to keep out large molecules, maintaining or determining shape, providing protection from attack, contributing to virulence, and enabling enhanced surface adhesion (31). In “Ca. Nitrososphaera viennensis,” the S-layer is the only detected outer envelope structure and has been suggested to act as a cell wall, forming a pseudoperiplasmic space outside the plasma membrane (22). It is likely that the S-layer in “Ca. Nitrosotenuis aquarius” also functions similarly to a cell wall, maintaining cell shape and structural integrity and associating closely with the cell membrane. Based on modeling data (32), the S-layer in “Ca. Nitrosotenuis aquarius” may also be important for concentrating ammonium to ammonia monooxygenase in the cytoplasmic membrane.
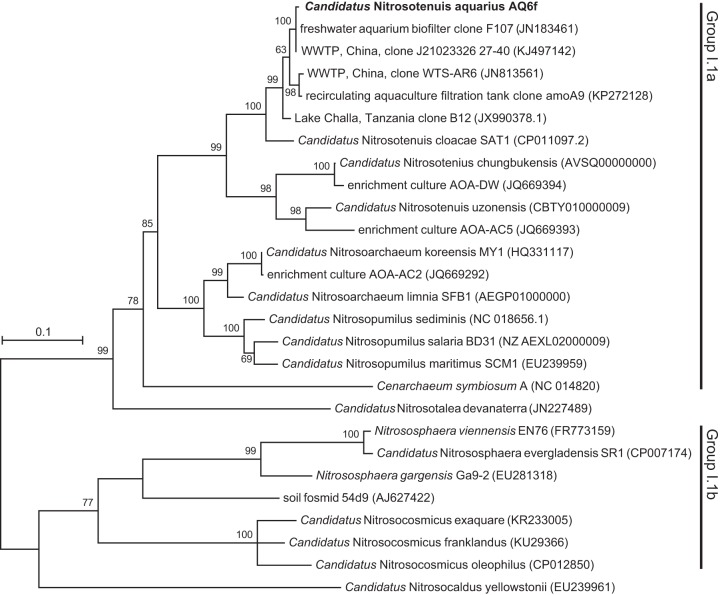
Based on amoA gene sequences, “Ca. Nitrosotenuis aquarius” clusters within the group I.1a Thaumarchaeota and belongs to the “Ca. Nitrosotenuis” lineage (Fig. 3). This lineage has high bootstrap support (99%) and contains previously described species, including “Ca. Nitrosotenuis uzonensis” (19), “Ca. Nitrosotenuis chungbukensis” (20), and “Ca. Nitrosotenuis cloacae” (24), as well as two AOA enrichment cultures without formal names, AOA-DW and AOA-AC2 (33). The placement of “Ca. Nitrosotenuis aquarius” within the amoA phylogeny was similar to that observed in a concatenated ribosomal tree with representative genomes (Fig. S2). The amoA gene of “Ca. Nitrosotenuis aquarius” is closely related to environmental homologues originating from freshwater aquarium biofilters (1), WWTPs, a recirculating aquaculture filtration tank (unpublished GenBank sequence), and Lake Challa (Tanzania) (34) (Fig. 3). The “Ca. Nitrosotenuis” clade does not contain any cultured marine AOA representatives, and this lineage appears to be characterized by low-salinity AOA.
FIG 3.
Phylogenetic relationship of “Ca. Nitrosotenuis aquarius” and cultivated AOA representatives based on amoA gene sequences. GenBank accession numbers for reference sequences are indicated in brackets. Bootstrap values are indicated above branches. Only bootstrap values greater than 50% are shown. The tree represents maximum likelihood based on the general time-reversible model. The scale bar represents 10% sequence divergence.
Based on available AOA genomes, the “Ca. Nitrosotenuis” lineage is characterized by higher G+C content than other representatives of group I.1a Thaumarchaeota (Table 1). Although genomes of the “Ca. Nitrosopumilus” and “Ca. Nitrosoarchaeum” lineages have <35% G+C content, available “Ca. Nitrosotenuis” genomes possess >41% G+C content. Additional genomes follow the same pattern, including “Ca. Nitrosotenuis chungbukensis” (42%) (20), “Candidatus Nitrosopelagicus brevis” (33%) (35), “Candidatus Nitrosopumilus salaria” (34%) (36), and “Candidatus Nitrosopumilus sediminis” (34%) (37).
TABLE 1.
Genome features of “Ca. Nitrosotenuis aquarius” and selected Thaumarchaeota representatives
| Genome feature | Value for the parametera |
|||||||
|---|---|---|---|---|---|---|---|---|
| “Ca. Nitrosotenuis aquarius” AQ6F | “Ca. Nitrosotenuis uzonensis” N4 | “Ca. Nitrosotenuis cloacae” SAT1 | “Ca. Nitrosoarchaeum limnia” BG20 | “Ca. Nitrosopumilus maritimus” SCM1 | “Ca. Nitrosotalea devanaterra” Nd1 | Nitrososphaera viennensis EN76 | “Ca. Nitrosocosmicus exaquare” G61 | |
| Cluster | I.1a | I.1a | I.1a | I.1a | I.1a | I.1a-associated | I.1b | I.1b |
| Habitat | Freshwater aquarium | Hot spring | WWTP | Estuary | Marine aquarium | Acidic soil | Garden soil | WWTP |
| Genome size (Mb) | 1.70 | 1.65 | 1.62 | 1.86 | 1.65 | 1.81 | 2.53 | 2.99 |
| GC content (%) | 42.2 | 42.3 | 41.0 | 32.5 | 34.2 | 37.1 | 52.7 | 33.9 |
| Total no. of genomic objects | 2,088 | 1,999 | 1,955 | 2,632 | 2,012 | 2,144 | 3,267 | 3,391 |
| No. of protein-coding genes | 2,047 | 1,958 | 1,911 | 2,589 | 1,976 | 2,106 | 3,128 | 3,354 |
| Coding density (%) | 92.2 | 90.4 | 91.9 | 87.2 | 91.7 | 90.6 | 86.4 | 77.1 |
| 5S rRNA copy no. | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 16S/23S rRNA copy no. | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| No. of tRNAs | 38 | 38 | 40 | 38 | 40 | 37 | 39 | 39 |
| amoA copy no. | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| amoB copy no. | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| amoC copy no. | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 |
| Nitrite reductase (nirK) | + | + | + | + | + | + | + | + |
| No. of ammonium transporters | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 1 |
| Urease and accessory proteins | + | − | + | − | − | − | + | + |
| Urea transporters | − | − | +c | − | − | − | + | + |
| Carbon fixation | 3HP/4HB | 3HP/4HB | 3HP/4HB | 3HP/4HB | 3HP/4HB | 3HP/4HB | 3HP/4HB | 3HP/4HB |
| Flagellar proteins | + | + | + | + | − | + | + | − |
| Chemotaxis proteins | + | + | + | + | − | + | + | − |
| S-layer-associated proteins | + | + | + | + | + | + | + | − |
| Coenzyme F420 | + | + | + | + | + | + | + | + |
| Cobalamin synthesis | + | + | + | + | + | + | + | + |
| Short-chain PHA synthesis (phaC or phaE)b | + | + | + | + | + | + | + | + |
| Dicarboxylate transporter (SdcS) | + | + | − | − | + | − | − | + |
| Citrate transporter | + | − | − | + | − | − | − | − |
| Amino acid/oligopeptide transport | + | + | + | + | + | + | + | + |
| Reference | This work | 19 | 24 | 17 | 102 | 44 | 22 | 39 |
Plus and minus signs indicate presence and absence, respectively.
PHA, polyhydroxyalkanoic acid.
Truncated transporter.
“Ca. Nitrosotenuis aquarius” genome.
The genome of “Ca. Nitrosotenuis aquarius” was assembled into one continuous sequence that was 1,697,207 bases in length (Fig. S3). “Ca. Nitrosotenuis aquarius” harbors several genes related to chemolithoautotrophic ammonia oxidation (Table 1), including the amoA, amoB, and amoC subunits of ammonia monooxygenase, a nitrite reductase (nirK), and ammonium transporters, as well as urease subunits (ureA, ureB, and ureC) and accessory proteins (ureD, ureE, and ureF). Urease genes and transporters are found in all group I.1b soil AOA genomes available (25, 38–40) and in some group I.1a genomes, including “Candidatus Cenarchaeum symbiosum” (40), “Ca. Nitrosopumilus sediminis” (41), and “Ca. Nitrosotenuis chungbukensis” (20). Urea transporter genes were not detected in the genome of “Ca. Nitrosotenuis aquarius” by automatic annotation or BLAST searches of urea transporter genes from other AOA genomes. Because small uncharged polar molecules, such as urea, can diffuse at low rates across lipid bilayers, it is possible that “Ca. Nitrosotenuis aquarius” can use urea even without active transport. However, the closely related “Ca. Nitrosotenuis cloacae” harbors urease genes but has a truncated urea transporter and does not show any nitrite production when incubated with urea in the absence of ammonia (24), suggesting that transport may be important for urea degradation. It remains unclear whether “Ca. Nitrosotenuis aquarius” can actively degrade urea under some circumstances. Given that the genome does not contain urea transporters and that nitrite production for growth on urea was much lower than for growth on ammonia (Fig. S1), our data suggest that urea is not a primary source of energy.
In addition to genes for chemolithoautotrophic growth (e.g., those of the 3-hydroxypropionate/4-hydroxybutyrate [3HP/4HB] cycle), the genome of “Ca. Nitrosotenuis aquarius” encodes multicopper oxidases, which are found in other AOA and have been proposed as candidates for an important function in hydroxylamine oxidation (42). Although the genome does not possess a cyanase gene, “Ca. Nitrosotenuis aquarius” harbors genes for several transporters or permeases of organic carbon, including dicarboxylic acids, citrate, taurine, glycerol, amino acids, oligopeptides, and polyamines (e.g., spermidine and putrescine). Similar transporters are found in other group I.1a Thaumarchaeota (5, 17), but to date there is no direct evidence of AOA assimilating or oxidizing any of these compounds.
The “Ca. Nitrosotenuis aquarius” genome contains a gene cluster encoding several flagellar proteins, including genes encoding archaeal flagellin and flagellar assembly and accessory proteins (Fig. S4 and Table S2). Immediately downstream of the flagellar gene cluster, there are several genes encoding chemotaxis-related proteins, suggesting that the motility of “Ca. Nitrosotenuis aquarius” is driven by chemical signals, which is supported by cellular appendages observed on cells (Fig. 2D and E). Several AOA encode chemotaxis and flagellar proteins, including all available Nitrososphaera genomes (22, 38, 43), “Ca. Nitrosotalea devanaterra” (44), and several group I.1a AOA (17, 19, 20, 24, 37, 45).
Polyamines are organic polycations containing multiple amine groups, such as spermidine, spermine, and putrescine (46). The “Ca. Nitrosotenuis aquarius” genome encodes synthesis of the polyamines spermine (NAQ_1931) and spermidine (NAQ_0236), and similar genes are present in “Candidatus Nitrososphaera gargensis,” “Ca. Nitrososphaera viennensis,” “Candidatus Nitrososphaera evergladensis,” and “Ca. Nitrosotenuis uzonensis.” All available AOA genomes encode putrescine biosynthesis from agmatinase (speB; NAQ_0259) and deoxyhypusine synthase (NAQ_1235 and NAQ_1236), which produces the amino acid hypusine from spermidine, suggesting a widespread role for polyamines in AOA. Polyamines can be involved in many biological processes, such as nucleic acid and protein synthesis, membrane stabilization, and enzyme stimulation (46–48), and can serve as sources of carbon, nitrogen, and energy to heterotrophic bacteria (49). Importantly, marine AOA enrichment cultures have been reported to oxidize putrescine and other polyamines directly to nitrite (50). “Ca. Nitrosotenuis aquarius” also encodes the PotABCD system for import of polyamines, an uptake system that has been identified in Escherichia coli, cyanobacteria, yeast, and several marine bacterial groups (51–53). Interestingly, genes encoding this system were not detected in any other AOA genomes. The ability to import exogenous polyamines could provide an advantage in reducing the energy expenditure needed to synthesize them or even provide an alternate source of energy.
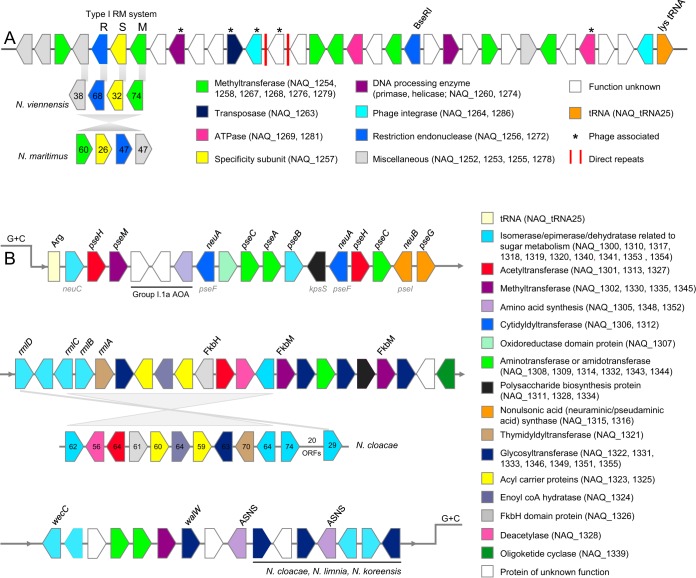
The genome of “Ca. Nitrosotenuis aquarius” contains two regions that have distinct G+C contents compared to content of the rest of the genome (regions R1 and R2 in Fig. S3). Region R1 consists of approximately 34 open reading frames (ORFs), several of which are related to restriction-modification systems, such as restriction endonucleases, components of a type I restriction-modification system (M subunit and S subunit), ATPases, and several DNA methyltransferases (Fig. 4A and Table S3). One cluster of four ORFs encoding a type I restriction-modification system is shared by “Ca. Nitrososphaera viennensis” and “Ca. Nitrosopumilus maritimus,” but none of the other genes in this segment share close identity to genes in any other AOA with sequenced genomes. In addition, “Ca. Nitrosotenuis aquarius” encodes BseR1, a type IIS restriction endonuclease discovered in Bacillus spp. (54). This DNA segment contains several hallmarks of genetic islands (55): it is flanked by a tRNA gene and contains integrase genes, direct repeats, and phage-related genes. Given these characteristics, its absence in related AOA, and the distinct nucleotide frequency, this segment of DNA likely arose from horizontal gene transfer (56, 57) and may have been maintained by “Ca. Nitrosotenuis aquarius” because it confers resistance to viral attacks via acquired restriction enzymes.
FIG 4.
Gene schematics of low-GC regions of the “Ca. Nitrosotenuis aquarius” genome. ORFs on forward and reverse strands are indicated by arrow directions and are not drawn to scale. Gene locus tags are indicated for assigned categories. (A) Defense-related genomic segment (R1 on Fig. S3 in the supplemental material). Numbers inside arrows represent amino acid identities (percent) to “Ca. Nitrosotenuis aquarius” proteins. (B) Glycosylation-related genomic segment (R2 on Fig. S3). Gene groups with lines below indicate that homologous genes in conserved order exist in the organisms indicated. Where available, specific gene names are indicated above the ORFs. Gene names indicated in gray below ORFs indicate potential gene alternatives. The largest conserved region within this DNA segment was a 12-ORF region in “Ca. Nitrosotenuis cloacae.” Numbers shown inside arrows represent percent amino acid identity to homologues in “Ca. Nitrosotenuis aquarius.” pse genes encode proteins involved in pseudaminic acid biosynthesis. The neu and kps genes encode proteins involved in neuraminic acid synthesis. Fkb proteins are involved in polyketide synthesis. rml genes are involved in rhamnose synthesis. Annotations for all genes in this figure are available in Tables S3 and S4.
“Ca. Nitrosotenuis aquarius” possesses a second region with distinct G+C content (region R2 in Fig. S3). This region begins with a tRNA gene and contains approximately 50 ORFs related to polyketide and rhamnose synthesis (see the supplemental material) and also to glycosylation (Fig. 4B and Table S4). With respect to glycosylation, this region contains several epimerase, isomerase, and dehydratase enzymes related to nucleotide-sugar metabolism, as well as several glycosyltransferases, methyltransferases, and nucleotidyltransferases. In particular, this segment may be involved in the synthesis of nonulsonic acids for protein glycosylation, most likely pseudaminic acid (encoded by pse genes) or sialic acid (N-acetyl-neuraminic acid, encoded by neu genes) (see the supplemental material for more detail). In addition to the genes located in this cluster, two trans-sialidase genes (NAQ_0881 and NAQ_0882) are also present, which could be involved in moving synthesized nonulsonic acids to their final destinations. Several bacterial surface proteins and flagellins are modified with various glycan groups, including derivatives of nonulsonic acids (58). For example, pseudaminic acid is a virulence factor and is added to the flagella of Helicobacter pylori and Campylobacter jejuni (59) and to the pili of Pseudomonas aeruginosa (60). Sialic acid is also used by some Gram-positive bacteria for glycosylation of outer spore coats (61) and for production of polysialic acid capsules (62). Synthesis of nonulsonic acids for S-layer glycosylation was previously suggested for “Ca. Nitrosotalea devanaterra” based on several genes involved in the production of nonulsonic acids located next to the major S-layer protein (44). Although identity and synteny of nonulsonic acid synthesis genes are low between “Ca. Nitrosotenuis aquarius” and “Ca. Nitrosotalea devanaterra,” it is likely that overall functions are similar. In “Ca. Nitrosotenuis aquarius” it is possible that either the S-layers or observed appendages are glycosylated with nonulsonic acids. Glycoproteins have a variety of biological functions, such as increasing protein stability and rigidity, acting as virulence factors, cellular signaling, and adhesion to surfaces (31, 63). Therefore, it is possible that glycosylation could contribute to the ability of “Ca. Nitrosotenuis aquarius” to adhere to surfaces, an adaptation that would confer an ecological advantage in the filter biofilm where it originates. The presence of glycosyltransferases and polysaccharide biosynthesis genes in genomic islands is a common phenomenon (64), also previously observed in AOA (e.g., “Ca. Nitrosopelagicus brevis”) (35), and is suggested to be part of an evolutionary arms race against phage.
Overall, the “Ca. Nitrosotenuis aquarius” genome displays a high degree of synteny with other group I.1a Thaumarchaeota, especially “Ca. Nitrosotenuis cloacae” and “Ca. Nitrosotenuis uzonensis” (Fig. S5A and B). Compared to “Ca. Nitrosotenuis cloacae,” there is an apparent inversion of a large proportion of the genome (Fig. S5B), and compared to the marine representative “Ca. Nitrosopumilus maritimus,” the majority of the genome runs antiparallel (Fig. S5C). Conversely, negligible genome synteny was observed with the group I.1b representative “Ca. Nitrososphaera viennensis” (Fig. S5D), which is consistent with a more distant phylogenetic relationship (Fig. 3 and S2). Average nucleotide identity (ANI) and average amino acid identity (AAI) values demonstrated that “Ca. Nitrosotenuis aquarius” is most closely related to “Ca. Nitrosotenuis cloacae” with ANI and AAI values of 79.1% and 82.7%, respectively (Table 2). The close evolutionary relationship of these archaea is also supported by phylogenetic analyses (Fig. 3 and S2). Both ANI and AAI values offer better resolution for species delineation than the 16S rRNA gene and show higher correlations with DNA-DNA hybridization values (65). Next to “Ca. Nitrosotenuis cloacae,” “Ca. Nitrosotenuis aquarius” shared the highest AAI with “Ca. Nitrosotenuis uzonensis” at 73.0%, followed by AAI values of approximately 63% with other group I.1a Thaumarchaeota (Table 2). Microorganisms with ANIs above 94 to 95% are considered to belong to the same species (65, 66). The 70% DNA-DNA reassociation threshold used as the major criterion for species delineation (67) corresponds to an AAI of 95 to 96% (68). Based on the species criteria for both ANI and AAI values, “Ca. Nitrosotenuis aquarius” represents a novel species within the “Ca. Nitrosotenuis” genus.
TABLE 2.
ANI and AAI values for “Ca. Nitrosotenuis aquarius” with group I.1a Thaumarchaeota
| Organism | ANI (%)a | AAI (%)b |
|---|---|---|
| “Ca. Nitrosotenuis cloacae” SAT1 | 79.1 | 82.7 |
| “Ca. Nitrosotenuis uzonensis” N4 | 75.2 | 73.0 |
| “Ca. Nitrosotenuis chungbukensis” MY2 | 75.3 | ND |
| “Ca. Nitrosopumilus sediminis” AR2 | 74.7 | 62.5 |
| “Ca. Nitrosopumilus maritimus” SCM1 | 74.6 | 62.8 |
| “Ca. Nitrosoarchaeum limnia” SFB1 | 75.1 | 62.4 |
| “Ca. Nitrosoarchaeum limnia” BG20 | 76.3 | 62.8 |
| “Ca. Nitrosoarchaeum koreensis” MY1 | 75.4 | 62.9 |
ANI, average nucleotide identity.
AAI, average amino acid identity; ND, not determined.
Ammonia-oxidizing activity of aquarium filter biomass.
It has been demonstrated that AOA outnumber AOB in freshwater aquarium biofilters and that AOB are often below detection limits in these environments (1, 2, 4). Several studies have linked nitrification with AOA detected in the environment (69–73), but others have warned that the presence of AOA, even in high abundance, does not indicate ammonia-oxidizing activity (74, 75). The activity and genome of “Ca. Nitrosotenuis aquarius” demonstrate chemolithoautotrophy, providing support for the role of AOA as nitrifiers in aquarium biofilters.
To assess AOA and AOB activity in situ, biofilm material from the freshwater aquarium biofilter where “Ca. Nitrosotenuis aquarius” originates was incubated with ammonia and differential inhibitors at two time points, including December 2014 and May 2016. Prior to this, differential inhibitors of nitrification were tested on “Ca. Nitrosotenuis aquarius” enrichment cultures, and PTIO (2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide) was inhibitory at low concentrations (Fig. S6), consistent with previous studies (16, 76, 77), whereas octyne was not inhibitory at the low concentrations (e.g., 10 μM) used previously for specific inhibition of AOB (78). When incubated in aquarium water supplemented with 0.5 mM NH4Cl, aquarium filter biomass depleted all ammonia within approximately 2 days in the absence of inhibitors (Fig. 5A), whereas killed biomass controls showed no activity during the incubation period. Strong inhibition of ammonia oxidation was observed in the presence of 200 μM PTIO. Quantitative PCR (qPCR) for genomic DNA extracted from the biofilm demonstrated that archaeal amoA genes comprised 99.5% of the total detectable amoA gene signal, whereas betaproteobacterial amoA genes comprised <1% (Table S5). In addition, denaturing gradient gel electrophoresis (DGGE) fingerprinting for thaumarchaeotal 16S rRNA genes indicated that the dominant thaumarchaeotal band in the aquarium biofilm fingerprint migrated to the same position as the band from “Ca. Nitrosotenuis aquarius” (Fig. S7). Subsequent sequencing of excised bands verified 100% nucleotide identity between the dominant biofilm band and the “Ca. Nitrosotenuis aquarius” sequence.
FIG 5.
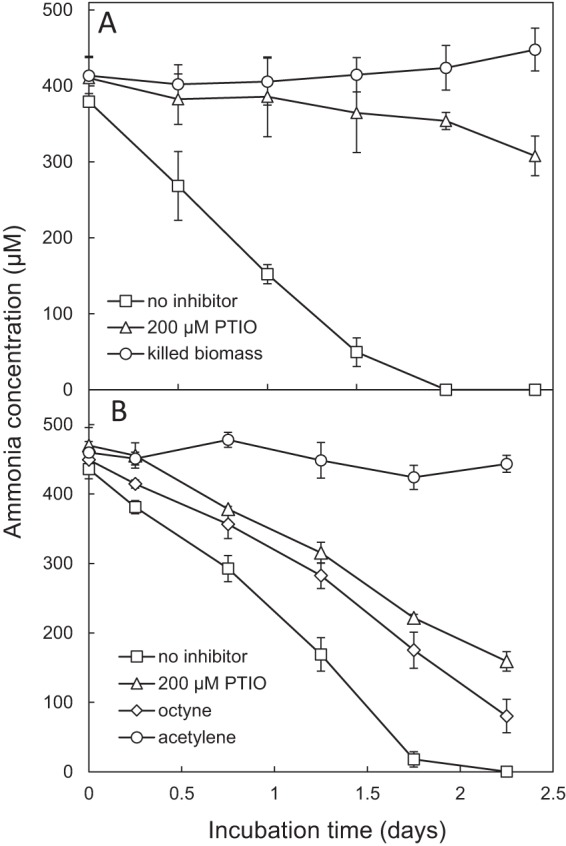
Ammonia-oxidizing activity of aquarium sponge filter biomass with nitrification inhibitors in an established aquarium biofilter (A) and in the same aquarium biofilter after it was decommissioned and reestablished (B). Error bars represent the standard errors of biological triplicates. Error bars not seen are contained within the symbols.
At the second time point, filter biomass was incubated with 0.5 mM NH4Cl and various differential inhibitors. Total depletion of ammonia occurred within approximately 2 days in the absence of inhibitors (Fig. 5B), similar to the results at the first time point. Addition of PTIO and octyne each resulted in partial inhibition of ammonia oxidation (Fig. 5B). When ammonia was depleted in the no-inhibitor control (1.75 days), ∼50% of the ammonia remained under the PTIO condition, whereas ∼40% of the ammonia remained in the octyne treatment. AOA contributed 92% of the total amoA signal at this time point (Table S5), and DGGE revealed only one thaumarchaeotal band, which corresponded to the “Ca. Nitrosotenuis aquarius” band (Fig. S7) and shared 100% sequencing identity. The increase in AOB-associated amoA genes (8%) (Table S5) compared to the level at the first time point is consistent with observed partial inhibition by octyne. These results suggest that AOA contribute to ammonia oxidation in freshwater aquaria but indicate that octyne-sensitive organisms may also contribute. It is possible that AOB contribute to ammonia-oxidizing activity in situ disproportionately to their abundance or, alternatively, that completely nitrifying (comammox) bacteria (79, 80), with yet-unknown inhibitor profiles, contributed to ammonia oxidation in these incubations.
Conclusions.
We report the cultivation and complete genome sequence of “Ca. Nitrosotenuis aquarius,” a mesophilic, autotrophic, ammonia-oxidizing archaeon that originates from a freshwater aquarium biofilter and belongs to the “Ca. Nitrosotenuis” lineage of the Thaumarchaeota. “Ca. Nitrosotenuis aquarius” cells grow up to 3 μm, which is the longest cell length reported for AOA representatives. Cells are consistently slender (0.4 μm), maintaining a high surface area-to-volume ratio, which is characteristic of oligotrophic cells. “Ca. Nitrosotenuis aquarius” possesses several genes for protein glycosylation, which could modify flagella or S-layer proteins, and may promote enhanced surface adhesion. Incubations of biomass from the freshwater aquarium biofilter where “Ca. Nitrosotenuis aquarius” originates demonstrate that AOA contribute to ammonia-oxidizing activity and support the previously reported numerical dominance of AOA in freshwater aquarium biofilters (1, 2). This work suggests that, contrary to common belief in the aquarium industry, AOB are likely not primarily responsible for ammonia oxidation in aquarium biofilters. Laboratory cultures of AOA originating from freshwater aquaria, such as “Ca. Nitrosotenuis aquarius” cultures, are useful for future investigations of the ecology and physiology of freshwater AOA, with potential commercial applications in aquarium and aquaculture operations.
MATERIALS AND METHODS
Cultivation.
Inoculum was obtained from a Fluval 404 Canister Filter sponge (Hagen, Baie d'Urfé, QC, Canada) from a previously described freshwater aquarium biofilter dominated by AOA (aquarium FW27) (1). To select for AOA, biomass was suspended in aquarium water and filtered through a 0.45-μm-pore-size syringe filter. Cultures were grown in medium used previously for AOB and AOA (81, 82), containing (per liter) 0.05 g of KH2PO4, 0.075 g of KCl, 0.05 g of MgSO4·7H2O, 0.58 g of NaCl, and 4 g of CaCO3. After autoclaving, the medium was supplemented with filter-sterilized NH4Cl (0.5 to 1 mM), 1 ml of selenite-tungstate solution, and 1 ml of trace element solution, as described previously (5, 83). The pH of the medium was approximately 8.5. Cultures were incubated at 28 to 30°C in the dark, without shaking. Enrichment of AOA was achieved using dilution to extinction and addition of antibiotics, including streptomycin (100 μg ml−1), ampicillin (100 μg ml−1), and nalidixic acid (30 μg ml−1).
Incubations for growth curve, temperature, ammonia inhibition, and growth with urea.
Incubations for growth curves were performed in quadruplicate 100-ml volumes, using the medium described above. Actively growing “Ca. Nitrosotenuis aquarius” cells were subcultured into fresh medium (1%, vol/vol, inoculum) and incubated at 30°C in the dark without shaking. Culture samples (2 ml) were removed every 2 days and pelleted for 10 min at 15,000 × g. Supernatants were removed for water chemistry measurements, and pellets were used for DNA extraction and quantitative PCR (qPCR).
To assess optimal growth temperature, “Ca. Nitrosotenuis aquarius” was incubated at a variety of temperatures. To assess ammonia tolerance, “Ca. Nitrosotenuis aquarius” was subcultured into fresh medium containing 0.25 to 10 mM ammonium chloride. To assess optimal salinity, “Ca. Nitrosotenuis aquarius” was incubated in 0 to 5% NaCl at 30°C. To assess the potential for growth on urea, “Ca. Nitrosotenuis aquarius” was grown in medium containing either 0.5 mM NH4Cl, 0.5 mM urea, or both 0.5 mM NH4Cl and 0.5 mM urea. Sterile controls containing 0.5 mM urea were included to assess the potential for ammonia production from spontaneous urea breakdown under the incubation conditions. In all cases, ammonia-oxidizing activity was assessed by nitrite production, and various conditions were set up on the same day, using the same inoculum and batch of growth medium. Unless otherwise indicated, all incubations were performed in triplicate using 1% subcultures in medium containing 0.5 mM ammonia and incubated at 30°C in the dark without shaking.
DNA extraction and quantitative PCR.
Genomic DNA extractions for growth curves were performed using an UltraClean Microbial DNA isolation kit (Mo Bio, Carlsbad, CA) according to the manufacturer's instructions, except that samples were incubated at 60°C prior to bead beating. All qPCR assays were performed in 10-μl reaction volumes containing 5 μl of 2× SsoAdvanced universal SYBR green supermix (Bio-Rad, Hercules, CA), 500 nM each primer, 5 μg of bovine serum albumin, and 0.1 to 5 ng of genomic DNA template. Primers used for amplification of thaumarchaeotal and bacterial 16S rRNA genes were the pair 771F and 957R (84) and the pair 341F and 518R (85), respectively. For thermal cycling details, see the supplemental material. For growth curves, the generation time of “Ca. Nitrosotenuis aquarius” was calculated using the slope of log-transformed thaumarchaeotal 16S rRNA gene copy numbers. Gene copy numbers were used as an approximation of cell numbers, assuming one 16S rRNA gene copy per genome and one chromosome per cell.
Phylogenetic analyses.
The full-length amoA gene sequence from “Ca. Nitrosotenuis aquarius” was compared to sequences of cultured AOA representatives and environmental clones. All reference sequences were obtained from GenBank, and nucleotide sequences were aligned with MUSCLE (86) in MEGA6 (87). Using the resulting alignment, an evolutionary history was inferred using the maximum likelihood method based on the general time-reversible model of sequence evolution, and the tree with the highest log likelihood was selected for display. A discrete gamma distribution was used to model evolutionary rate differences among sites (using five discrete gamma categories), and the rate variation model allowed for some sites to be evolutionarily invariable. The tree was then drawn to scale, with branch lengths measured in the number of substitutions per site. Bootstrap values were calculated based on 1,000 replicates and are indicated above tree branches. An additional phylogeny based on concatenated ribosomal proteins was also prepared using methods outlined in the supplemental material.
CARD-FISH.
For catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH), all samples were fixed with 4% formaldehyde at room temperature for 3 h and subjected to a standard ethanol series (50%, 80%, and 98%; 3 min each). CARD-FISH was performed as described previously (74, 88), with modifications. Briefly, fixed samples were applied to poly-l-lysine-coated slides and embedded in 0.2% agarose. Cells were permeabilized with 0.1 M HCl at room temperature for 30 s. To inactivate endogenous peroxidases, slides were treated with 3% H2O2 at room temperature for 10 min. Probe Arch915 (89) was labeled with horseradish peroxidase (HRP) and applied at a concentration of 0.17 ng μl−1. Probes were hybridized in a buffer containing 20% formamide for 2.5 h at 46°C and then washed in wash buffer (with corresponding stringency) for 15 min at 48°C. Fluorescein isothiocyanate (FITC) tyramides and H2O2 (0.15%) were added to the amplification buffer and incubated at 46°C for 30 min. All samples were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; 5 μg ml−1). In all cases, no-probe controls and HRP-labeled nonsense probes were prepared and imaged to ensure signal specificity.
Citifluor AF1 antifade mounting solution (Citifluor, London, United Kingdom) was applied to samples prior to imaging, and samples were visualized on an inverted Leica TCS SP8 confocal laser scanning microscope (CLSM). For imaging FITC signals, excitation and emission wavelengths were 492 nm and 520 nm, respectively. For imaging DAPI signal, excitation and emission wavelengths were 400 nm and 600 nm, respectively.
SEM and transmission electron microscopy.
For scanning electron microscopy (SEM), a 10-μl sample obtained from an actively growing culture was applied to a 5- by 5-mm silicon wafer (SPI Supplies, West Chester, PA) that was attached to an aluminum stub (VWR, Radnor, PA) with conductive carbon tape (VWR). Immediately after samples were dried in a fume hood, cells were imaged using a LEO 1550 field emission scanning electron microscope (Zeiss, Oberkochen, Germany). An InLens SE detector was used for high-resolution topographic imaging, with an electron high-voltage (EHT) setting of 5.00 to 7.00 kV and a working distance of 10.3 to 10.5 mm.
For transmission electron microscopy (TEM), cells from actively growing cultures were imaged without fixation. A 5-μl sample was placed onto a 200-mesh copper grid with Formvar/carbon coating (Electron Microscopy Sciences, Hatfield, PA). Samples were dried, washed with 0.05 M HEPES buffer to remove medium salts, and then negatively stained with 2% uranyl acetate. Imaging was performed using a Tecnai G2 F20 transmission electron microscope (FEI, Hillsboro, OR) operating at 200 kV with a bottom-mount Gatan 4K charge-coupled-device (CCD) camera and Digital Micrograph software (Gatan, Inc., Pleasanton, CA).
Genome sequencing and assembly.
Genomic DNA for metagenomic sequencing was extracted from “Ca. Nitrosotenuis aquarius” enrichment culture using a PowerSoil DNA isolation kit (Mo Bio Laboratories) according to the manufacturer's instructions. Enrichment culture samples (6 at 50 ml each) were pelleted at 7,200 × g for 30 min, supernatant was removed and discarded, and pellets were suspended in bead lysis solution for DNA extraction.
A short-insert, paired-end library was constructed using a NEBNext Ultra DNA library kit and sequenced with a single index using the Illumina platform (90) on a MiSeq (Illumina, San Diego, CA). In total, 26,430,800 reads of 251 bases in length were generated for a total of 6,634 Mbp. The data were quality trimmed using FaQCs (91) and assembled with SPAdes, version 3.5.0 (92). The assembled contigs were binned using MaxBin, version 1.4.5 (93), to isolate contigs for the genome of interest. Four contigs (contained in one bin) were identified as “Ca. Nitrosopumilus”-like using EDGE (94). These four contigs were extended computationally with the quality-trimmed data using PRICE (95), which closed the gap between two of the contigs, leaving three remaining contigs. Following assembly of Illumina reads, primer walking was used to close remaining gaps. Primers were designed from the ends of the extended contigs (see Table S1 in the supplemental material), and PCR and Sanger sequencing were used to close gaps. Primer design and assembly of overlapping Sanger sequences were performed in Geneious R9 (Biomatter, Ltd., Auckland, New Zealand).
Genome annotation and analysis.
Genome annotation was completed using two automatic annotation pipelines, including Integrated Microbial Genome/Expert Review (IMG/ER) (96) and the MicroScope Microbial Genome Annotation & Analysis Platform (MaGe) (101). For genes and regions of interest, manual curation was performed using MaGe. Unless otherwise indicated, locus tags and gene arrangements are based on MaGe annotations.
The average nucleotide identity (ANI) and average amino acid identity (AAI) of protein-coding genes were calculated using online tools (http://enve-omics.ce.gatech.edu/ani/), implementing methods described previously (65, 66, 97). For ANI values, a window size of 1,000 bases with a 200-base step size was used to determine reciprocal best hits (with blastn). A minimum alignment length of 700 bp was used with a minimum identity of 70%. For AAIs, protein-coding sequences were based on MaGe protein-coding gene predictions and were calculated based on reciprocal best hits (blastp) with a minimum alignment of 50% and a minimum identity of 20%. To determine genome synteny, full-genome alignments and dot plots between “Ca. Nitrosotenuis aquarius” and other AOA genomes were generated using PROmer (98) as implemented in IMG/ER. Six-frame amino acid translations of input DNA sequences were used to identify matches, and a match length of six amino acids was used.
Incubations of sponge filter with nitrification inhibitors.
To assess in situ contributions to ammonia oxidation, biofilm from the sponge filter from which “Ca. Nitrosotenuis aquarius” was enriched (aquarium FW27) were incubated with differential inhibitors at two time points (December 2014 and April 2016). At the first time point, December 2014, the aquarium had been established for approximately 6 years; but the fish and filter were then moved to a different location, and the aquarium was dismantled and out of commission for approximately 10 months. The aquarium was reestablished using the same fish and filter system for approximately 8 months prior to the second time point. For these incubations, biofilm was squeezed from the sponge filter into aquarium water, resulting in a slurry that was distributed in 25-ml volumes to 125-ml serum bottles with silicone stoppers. All flasks were amended with 0.5 mM NH4Cl and incubated at 28°C in the dark, and all incubations were performed in triplicate. At the first time point, conditions included no-inhibitor controls, 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide (PTIO; 200 μM), and paraformaldehyde (PFA)-killed biomass controls. At the second time point, conditions included no-inhibitor controls, PTIO (200 μM), octyne (6 μM; aqueous), and acetylene (4 μM; aqueous).
Water chemistry measurements.
Nitrite and ammonia measurements were performed using colorimetric protocols as reported previously (99, 100). For both nitrite and ammonia, absorbance was measured with a FilterMax F5 multimode plate reader (Molecular Devices) at 550 nm and 450 nm, respectively.
Accession number(s).
The full genome sequence of “Ca. Nitrosotenuis aquarius” has been deposited in GenBank under accession number CP024808, and associated annotations are available in IMG/ER (identification number 78086) and MaGe (T3JK6F).
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael Wagner for access to microscopy facilities and Robert Harris and Brandon Seo for technical assistance with electron microscopy. We thank Barbara Butler for helpful suggestions. We acknowledge assistance from Olga Anna Duhl and John C. Semple during the process of selecting a Latin species name for “Candidatus Nitrosotenuis aquarius.”
This research was funded by a Vanier Canada Graduate Scholarship to L.A.S. and a Discovery Grant to J.D.N., both provided by the Natural Sciences and Engineering Council of Canada.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01430-18.
REFERENCES
- 1.Sauder LA, Engel K, Stearns JC, Masella AP, Pawliszyn R, Neufeld JD. 2011. Aquarium nitrification revisited: Thaumarchaeota are the dominant ammonia oxidizers in freshwater aquarium biofilters. PLoS One 6:e23281. doi: 10.1371/journal.pone.0023281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagchi S, Vlaeminck SE, Sauder LA, Mosquera M, Neufeld JD, Boon N. 2014. Temporal and spatial stability of ammonia-oxidizing archaea and bacteria in aquarium biofilters. PLoS One 9:e113515. doi: 10.1371/journal.pone.0113515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrell PC, Phalen CM, Hovanec TA. 2001. Identification of bacteria responsible for ammonia oxidation in freshwater aquaria. Appl Environ Microbiol 67:5791–5800. doi: 10.1128/AEM.67.12.5791-5800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hovanec TA, DeLong EF. 1996. Comparative analysis of nitrifying bacteria associated with freshwater and marine aquaria. Appl Environ Microbiol 62:2888–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 6.Urakawa H, Tajima Y, Numata Y, Tsuneda S. 2008. Low temperature decreases the phylogenetic diversity of ammonia-oxidizing archaea and bacteria in aquarium biofiltration systems. Appl Environ Microbiol 74:894–900. doi: 10.1128/AEM.01529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakami T, Andoh T, Morita T, Yamamoto Y. 2012. Phylogenetic diversity of ammonia-oxidizing archaea and bacteria in biofilters of recirculating aquaculture systems. Mar Genomics 7:27–31. doi: 10.1016/j.margen.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Srithep P, Khinthong B, Chodanon T, Powtongsook S, Pungrasmi W, Limpiyakorn T. 2015. Communities of ammonia-oxidizing bacteria, ammonia-oxidizing archaea and nitrite-oxidizing bacteria in shrimp ponds. Ann Microbiol 65:267–278. doi: 10.1007/s13213-014-0858-3. [DOI] [Google Scholar]
- 9.Brown MN, Briones A, Diana J, Raskin L. 2013. Ammonia-oxidizing archaea and nitrite-oxidizing nitrospiras in the biofilter of a shrimp recirculating aquaculture system. FEMS Microbiol Ecol 83:17–25. doi: 10.1111/j.1574-6941.2012.01448.x. [DOI] [PubMed] [Google Scholar]
- 10.Martens-Habbena W, Stahl DA. 2011. Nitrogen metabolism and kinetics of ammonia-oxidizing archaea. Methods Enzymol 496:465–487. doi: 10.1016/B978-0-12-386489-5.00019-1. [DOI] [PubMed] [Google Scholar]
- 11.Merbt SN, Stahl DA, Casamayor EO, Martí E, Nicol GW, Prosser JI. 2012. Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiol Lett 327:41–46. doi: 10.1111/j.1574-6968.2011.02457.x. [DOI] [PubMed] [Google Scholar]
- 12.Busiek KK, Margolin W. 2011. Split decision: a thaumarchaeon encoding both FtsZ and Cdv cell division proteins chooses Cdv for cytokinesis. Mol Microbiol 82:535–538. doi: 10.1111/j.1365-2958.2011.07833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelve EA, Lindås A-C, Martens-Habbena W, de la Torre JR, Stahl DA, Bernander R. 2011. Cdv-based cell division and cell cycle organization in the thaumarchaeon Nitrosopumilus maritimus. Mol Microbiol 82:555–566. doi: 10.1111/j.1365-2958.2011.07834.x. [DOI] [PubMed] [Google Scholar]
- 14.Ng K-H, Srinivas V, Srinivasan R, Balasubramanian M. 2013. The Nitrosopumilus maritimus CdvB, but not FtsZ, assembles into polymers. Archaea 2013:104147. doi: 10.1155/2013/104147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vajrala N, Martens-Habbena W, Sayavedra-Soto LA, Schauer A, Bottomley PJ, Stahl DA, Arp DJ. 2013. Hydroxylamine as an intermediate in ammonia oxidation by globally abundant marine archaea. Proc Natl Acad Sci U S A 110:1006–1011. doi: 10.1073/pnas.1214272110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martens-Habbena W, Qin W, Horak REA, Urakawa H, Schauer AJ, Moffett JW, Armbrust EV, Ingalls AE, Devol AH, Stahl DA. 2015. The production of nitric oxide by marine ammonia-oxidizing archaea and inhibition of archaeal ammonia oxidation by a nitric oxide scavenger. Environ Microbiol 17:2261–2274. doi: 10.1111/1462-2920.12677. [DOI] [PubMed] [Google Scholar]
- 17.Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. 2011. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosier AC, Lund MB, Francis CA. 2012. Ecophysiology of an ammonia-oxidizing archaeon adapted to low-salinity habitats. Microb Ecol 64:955–963. doi: 10.1007/s00248-012-0075-1. [DOI] [PubMed] [Google Scholar]
- 19.Lebedeva E, Hatzenpichler R, Pelletier E, Schuster N, Hauzmayer S, Bulaev A, Grigor'eva NV, Galushko A, Schmid M, Palatinszky M, Le Paslier D, Daims H, Wagner M. 2013. Enrichment and genome sequence of the group I.1a ammonia-oxidizing archaeon “Ca. Nitrosotenuis uzonensis” representing a clade globally distributed in thermal habitats. PLoS One 8:e80835. doi: 10.1371/journal.pone.0080835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung M-Y, Park S-J, Kim S-J, Kim J-G, Sinninghe Damsté JS, Jeon CO, Rhee S-K. 2014. A mesophilic, autotrophic, ammonia-oxidizing archaeon of thaumarchaeal group I.1a cultivated from a deep oligotrophic soil horizon. Appl Environ Microbiol 80:3645–3655. doi: 10.1128/AEM.03730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA. 2008. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol 10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 22.Stieglmeier M, Klingl A, Alves RJ, Rittmann SK, Melcher M, Leisch N, Schleper C. 2014. Nitrososphaera viennensis gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int J Syst Evol Microbiol 64:2738–2752. doi: 10.1099/ijs.0.063172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. 2011. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci U S A 108:15892–15897. doi: 10.1073/pnas.1107196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Ding K, Wen X, Zhang B, Shen B, Yang Y. 2016. A novel ammonia-oxidizing archaeon from wastewater treatment plant: its enrichment, physiological and genomic characteristics. Sci Rep 6:23747. doi: 10.1038/srep23747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung M-Y, Park S-J, Min D, Kim J-S, Rijpstra WIC, Sinninghe Damste JS, Kim G-J, Madsen EL, Rhee S-K. 2011. Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group I.1a from an agricultural soil. Appl Environ Microbiol 77:8635–8647. doi: 10.1128/AEM.05787-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood CM. 1993. Ammonia and urea metabolism and excretion, p 381–425. In Evans DH. (ed), The physiology of fishes. CRC Press, Boca Raton, FL. [Google Scholar]
- 27.Walsh P, Milligan C. 1995. Effects of feeding and confinement on nitrogen metabolism and excretion in the gulf toadfish Opsanus beta. J Exp Biol 198:1559–1566. [DOI] [PubMed] [Google Scholar]
- 28.Satoh Y. 1980. Production of urea by bacterial decomposition of organic matter including phytoplankton. Int Rev Hydrobiol 65:295–301. doi: 10.1002/iroh.19800650216. [DOI] [Google Scholar]
- 29.Pedersen H, Lomstein BA, Henry BT. 1993. Evidence for bacterial urea production in marine sediments. FEMS Microbiol Ecol 12:51–59. doi: 10.1111/j.1574-6941.1993.tb00016.x. [DOI] [Google Scholar]
- 30.Hatzenpichler R. 2012. Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl Environ Microbiol 78:7501–7510. doi: 10.1128/AEM.01960-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sára M, Sleytr UB. 2000. S-layer proteins. J Bacteriol 182:859–868. doi: 10.1128/JB.182.4.859-868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li P-N, Herrmann J, Tolar BB, Poitevin F, Ramdasi R, Bargar JR, Stahl DA, Jensen GJ, Francis CA, Wakatsuki S, van den Bedem H. 13 June 2018. Nutrient transport suggests an evolutionary basis for charged archaeal surface layer proteins. ISME J doi: 10.1038/s41396-018-0191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.French E, Kozlowski JA, Mukherjee M, Bullerjahn G, Bollmann A. 2012. Ecophysiological characterization of ammonia-oxidizing archaea and bacteria from freshwater. Appl Environ Microbiol 78:5773–5780. doi: 10.1128/AEM.00432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buckles LK, Villanueva L, Weijers JWH, Verschuren D, Damsté JSS. 2013. Linking isoprenoidal GDGT membrane lipid distributions with gene abundances of ammonia-oxidizing Thaumarchaeota and uncultured crenarchaeotal groups in the water column of a tropical lake (Lake Challa, East Africa). Environ Microbiol 15:2445–2462. doi: 10.1111/1462-2920.12118. [DOI] [PubMed] [Google Scholar]
- 35.Santoro AE, Dupont CL, Richter RA, Craig MT, Carini P, McIlvin MR, Yang Y, Orsi WD, Moran DM, Saito MA. 2015. Genomic and proteomic characterization of “Candidatus Nitrosopelagicus brevis”: an ammonia-oxidizing archaeon from the open ocean. Proc Natl Acad Sci U S A 112:1173–1178. doi: 10.1073/pnas.1416223112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mosier AC, Allen EE, Kim M, Ferriera S, Francis CA. 2012. Genome sequence of “Candidatus Nitrosopumilus salaria” BD31, an ammonia-oxidizing archaeon from the San Francisco Bay estuary. J Bacteriol 194:2121–2122. doi: 10.1128/JB.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park S-J, Kim J-G, Jung M-Y, Kim S-J, Cha I-T, Ghai R, Martín-Cuadrado A-B, Rodríguez-Valera F, Rhee S-K. 2012. Draft genome sequence of an ammonia-oxidizing archaeon, “Candidatus Nitrosopumilus sediminis” AR2, from Svalbard in the Arctic Circle. J Bacteriol 194:6948–6949. doi: 10.1128/JB.01869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhalnina K, Dias R, Leonard MT, Dorr de Quadros P, Camargo FAO, Drew JC, Farmerie WG, Daroub SH, Triplett EW. 2014. Genome sequence of Candidatus Nitrososphaera evergladensis from group I.1b enriched from everglades soil reveals novel genomic features of the ammonia-oxidizing archaea. PLoS One 9:e101648. doi: 10.1371/journal.pone.0101648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sauder LA, Albertsen M, Engel K, Schwarz J, Nielsen PH, Wagner M, Neufeld JD. 2017. Cultivation and characterization of Candidatus Nitrosocosmicus exaquare, an ammonia-oxidizing archaeon from a municipal wastewater treatment plant. ISME J 11:1142–1157. doi: 10.1038/ismej.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, DeLong EF. 2006. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park S-J, Kim J-G, Jung M-Y, Kim S-J, Cha I-T, Kwon K, Lee J-H, Rhee S-K. 2012. Draft genome sequence of an ammonia-oxidizing archaeon, “Candidatus Nitrosopumilus koreensis” AR1, from marine sediment. J Bacteriol 194:6940–1941. doi: 10.1128/JB.01857-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozlowski JA, Stieglmeier M, Schleper C, Klotz MG, Stein LY. 2016. Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota. ISME J 10:1836–1845. doi: 10.1038/ismej.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spang A, Poehlein A, Offre P, Zumbrägel S, Haider S, Rychlik N, Nowka B, Schmeisser C, Lebedeva E, Rattei T, Böhm C, Schmid M, Galushko A, Hatzenpichler R, Weinmaier T, Daniel R, Schleper C, Spieck E, Streit W, Wagner M. 2012. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol 14:3122–3145. doi: 10.1111/j.1462-2920.2012.02893.x. [DOI] [PubMed] [Google Scholar]
- 44.Lehtovirta-Morley LE, Sayavedra-Soto LA, Gallois N, Schouten S, Stein LY, Prosser JI, Nicol GW. 2016. Identifying potential mechanisms enabling acidophily in the ammonia-oxidizing archaeon “Candidatus Nitrosotalea devanaterra.” Appl Environ Microbiol 82:2608–2619. doi: 10.1128/AEM.04031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim BK, Jung M-Y, Yu DS, Park S-J, Oh TK, Rhee S-K, Kim JF. 2011. Genome sequence of an ammonia-oxidizing soil archaeon, “Candidatus Nitrosoarchaeum koreensis” MY1. J Bacteriol 193:5539–5540. doi: 10.1128/JB.05717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tabor CW, Tabor H. 1985. Polyamines in microorganisms. Microbiol Rev 49:81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abraham KA. 1968. Studies on DNA-dependent RNA polymerase from Escherichia coli. The mechanism of polyamine induced stimulation of enzyme activity. Eur J Biochem 5:143–146. [DOI] [PubMed] [Google Scholar]
- 48.Frydman L, Rossomando PC, Frydman V, Fernandez C, Frydman B, Samejimat K. 1992. Interactions between natural polyamines and tRNA: an 15N NMR analysis. Proc Natl Acad Sci U S A 89:9186–9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Höfle MG. 1984. Degradation of putrescine and cadaverine in seawater cultures by marine bacteria. Appl Environ Microbiol 47:843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hollibaugh JT, Tolar BB, Popp BB, Wallsgrove NJ. 2015. Evidence for the direct oxidation of polyamine nitrogen by Thaumarchaeota-dominated marine nitrifying communities, abstr 26435. Abstr 2015 Aquat Sci Meet. Association for the Sciences of Limnology and Oceanography, Waco, TX. [Google Scholar]
- 51.Incharoensakdi A, Jantaro S, Raksajit W, Mäenpää P. 2010. Polyamines in cyanobacteria: biosynthesis, transport and abiotic stress response, p 23–32. In Mendez-Vilas A (ed), Current research, technology and education topics in applied microbiology and microbial biotechnology. Formatex, Badajoz, Spain. [Google Scholar]
- 52.Igarashi K, Kashiwagi K. 1999. Polyamine transport in bacteria and yeast. Biochem J 344:633–642. doi: 10.1042/bj3440633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mou X, Sun S, Rayapati P, Moran M. 2010. Genes for transport and metabolism of spermidine in Ruegeria pomeroyi DSS-3 and other marine bacteria. Aquat Microb Ecol 58:311–321. doi: 10.3354/ame01367. [DOI] [Google Scholar]
- 54.Mushtaq R, Naeem S, Sohail A, Riazuddin S. 1993. BseRI a novel restriction endonuclease from a Bacillus species which recognizes the sequence 5′… GAGGAG… 3′. Nucleic Acids Res 21:3585. doi: 10.1093/nar/21.15.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Juhas M, Van Der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawrence JG, Ochman H. 2002. Reconciling the many faces of lateral gene transfer. Trends Microbiol 10:1–4. doi: 10.1016/S0966-842X(01)02282-X. [DOI] [PubMed] [Google Scholar]
- 57.Ravenhall M, Škunca N, Lassalle F, Dessimoz C. 2015. Inferring horizontal gene transfer. PLoS Comput Biol 11:e1004095. doi: 10.1371/journal.pcbi.1004095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nothaft H, Szymanski CM. 2010. Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol 8:765–778. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- 59.Thibault P, Logan SM, Kelly JF, Brisson JR, Ewing CP, Trust TJ, Guerry P. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J Biol Chem 276:34862–34870. doi: 10.1074/jbc.M104529200. [DOI] [PubMed] [Google Scholar]
- 60.Castric P, Cassels FJ, Carlson RW. 2001. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J Biol Chem 276:26479–26485. doi: 10.1074/jbc.M102685200. [DOI] [PubMed] [Google Scholar]
- 61.Bliss JM, Silver RP. 1996. Coating the surface: a model for expression of capsular polysialic acid in Escherichia coli K1. Mol Microbiol 21:221–231. doi: 10.1046/j.1365-2958.1996.6461357.x. [DOI] [PubMed] [Google Scholar]
- 62.Driks A. 1999. Bacillus subtilis spore coat. Microbiol Mol Biol Rev 63:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Larkin A, Imperiali B. 2011. The expanding horizons of asparagine-linked glycosylation. Biochemistry 50:4411–4426. doi: 10.1021/bi200346n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodriguez-Valera F, Martin-Cuadrado A-B, Rodriguez-Brito B, Pašić L, Thingstad TF, Rohwer F, Mira A. 2009. Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 65.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A 102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 67.Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Truper HG. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol 37:463–464. doi: 10.1099/00207713-37-4-463. [DOI] [Google Scholar]
- 68.Konstantinidis KT, Tiedje JM. 2005. Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GJ, Middelburg JJ, Schouten S, Sinninghe Damsté JS. 2006. Archaeal nitrification in the ocean. Proc Natl Acad Sci U S A 103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gubry-Rangin C, Nicol GW, Prosser JI. 2010. Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol 74:566–574. doi: 10.1111/j.1574-6941.2010.00971.x. [DOI] [PubMed] [Google Scholar]
- 71.Zhang L-M, Hu H-W, Shen J-P, He J-Z. 2012. Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6:1032–1045. doi: 10.1038/ismej.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan J, Haaijer SCM, Op den Camp HJM, van Niftrik L, Stahl DA, Könneke M, Rush D, Sinninghe Damsté JS, Hu YY, Jetten MSM. 2012. Mimicking the oxygen minimum zones: stimulating interaction of aerobic archaeal and anaerobic bacterial ammonia oxidizers in a laboratory-scale model system. Environ Microbiol 14:3146–3158. doi: 10.1111/j.1462-2920.2012.02894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alves RJE, Wanek W, Zappe A, Richter A, Svenning MM, Schleper C, Urich T. 2013. Nitrification rates in Arctic soils are associated with functionally distinct populations of ammonia-oxidizing archaea. ISME J 7:1620–1631. doi: 10.1038/ismej.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mussmann M, Brito I, Pitcher A, Sinninghe Damste JS, Hatzenpichler R, Richter A, Nielsen JL, Nielsen PH, Muller A, Daims H, Wagner M, Head IM. 2011. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc Natl Acad Sci U S A 108:16771–16776. doi: 10.1073/pnas.1106427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sterngren AE, Hallin S, Bengtson P. 2015. Archaeal ammonia oxidizers dominate in numbers, but bacteria drive gross nitrification in N-amended grassland soil. Front Microbiol 6:1350. doi: 10.3389/fmicb.2015.01350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen T, Stieglmeier M, Dai J, Urich T, Schleper C. 2013. Responses of the terrestrial ammonia-oxidizing archaeon Ca. Nitrososphaera viennensis and the ammonia-oxidizing bacterium Nitrosospira multiformis to nitrification inhibitors. FEMS Microbiol Lett 344:121–129. doi: 10.1111/1574-6968.12164. [DOI] [PubMed] [Google Scholar]
- 77.Sauder LA, Ross AA, Neufeld JD. 2016. Nitric oxide scavengers differentially inhibit ammonia oxidation in ammonia-oxidizing archaea and bacteria. FEMS Microbiol Lett 363:fnw052. doi: 10.1093/femsle/fnw052. [DOI] [PubMed] [Google Scholar]
- 78.Taylor AE, Vajrala N, Giguere AT, Gitelman AI, Arp DJ, Myrold DD, Sayavedra-Soto L, Bottomley PJ. 2013. Use of aliphatic n-alkynes to discriminate soil nitrification activities of ammonia-oxidizing thaumarchaea and bacteria. Appl Environ Microbiol 79:6544–6551. doi: 10.1128/AEM.01928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Kessel MA, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJ, Kartal B, Jetten MS, Lücker S. 2015. Complete nitrification by a single microorganism. Nature 528:555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Daims H, Lebedeva E, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. 2015. Complete nitrification by Nitrospira bacteria. Nature 528:504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krümmel A, Harms H. 1982. Effect of organic matter on growth and cell yield of ammonia-oxidizing bacteria. Arch Microbiol 133:50–54. doi: 10.1007/BF00943769. [DOI] [Google Scholar]
- 82.Lebedeva E, Alawi M, Fiencke C, Namsaraev B, Bock E, Spieck E. 2005. Moderately thermophilic nitrifying bacteria from a hot spring of the Baikal rift zone. FEMS Microbiol Ecol 54:297–306. doi: 10.1016/j.femsec.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 83.Widdel F, Bak F. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p 3352–3378. In Balows A, Tr̈uper HG, Dworkin M, Harder W, Schleifer K-H (ed), The prokaryotes, 2nd ed Springer, New York, NY. [Google Scholar]
- 84.Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C. 2003. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol 5:787–797. doi: 10.1046/j.1462-2920.2003.00476.x. [DOI] [PubMed] [Google Scholar]
- 85.Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ishii K, Mussmann M, MacGregor BJ, Amann R. 2004. An improved fluorescence in situ hybridization protocol for the identification of bacteria and archaea in marine sediments. FEMS Microbiol Ecol 50:203–213. doi: 10.1016/j.femsec.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 89.Stahl DA, Amann R. 1991. Development and application of nucleic acid probes in bacterial systematics, p 205–248. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd, Chichester, England. [Google Scholar]
- 90.Bennett S. 2004. Solexa Ltd. Pharmacogenomics 5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 91.Lo C-C, Chain PSG. 2014. Rapid evaluation and quality control of next generation sequencing data with FaQCs. BMC Bioinformatics 15:366. doi: 10.1186/s12859-014-0366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu Y-W, Tang Y-H, Tringe SG, Simmons BA, Singer SW. 2014. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2:26. doi: 10.1186/2049-2618-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li P-E, Lo C-C, Anderson JJ, Davenport KW, Bishop-Lilly KA, Xu Y, Ahmed S, Feng S, Mokashi VP, Chain PSG. 2017. Enabling the democratization of the genomics revolution with a fully integrated web-based bioinformatics platform. Nucleic Acids Res 45:67–80. doi: 10.1093/nar/gkw1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ruby JG, Bellare P, DeRisi JL. 2013. PRICE: software for the targeted assembly of components of (meta)genomic sequence data. G3 (Bethesda) 3:865–880. doi: 10.1534/g3.113.005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Markowitz VM, Mavromatis K, Ivanova NN, Chen I-MA, Chu K, Kyrpides NC. 2009. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 97.Rodriguez LM, Konstantinidis KT. 2016. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr 4:e1900v1. [Google Scholar]
- 98.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Miranda KM, Espey MG, Wink DA. 2001. A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5:62–71. doi: 10.1006/niox.2000.0319. [DOI] [PubMed] [Google Scholar]
- 100.Meseguer-Lloret S, Molins-Legua C, Campins-Falco P. 2002. Ammonium determination in water samples by using OPA-NAC reagent: a comparative study with Nessler and ammonium selective electrode methods. Int J Environ Anal Chem 82:475–489. doi: 10.1080/0306731021000018107. [DOI] [Google Scholar]
- 101.Vallenet D, Engelen S, Mornico D, Cruveiller S, Fleury L, Lajus A, Rouy Z, Roche D, Salvignol G, Scarpelli C, Médigue C. 2009. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford) 2009:bap021. doi: 10.1093/database/bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, Brochier-Armanet C, Chain PSG, Chan PP, Gollabgir A, Hemp J, Hügler M, Karr EA, Könneke M, Shin M, Lawton TJ, Lowe T, Martens-Habbena W, Sayavedra-Soto LA, Lang D, Sievert SM, Rosenzweig AC, Manning G, Stahl DA. 2010. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci U S A 107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.