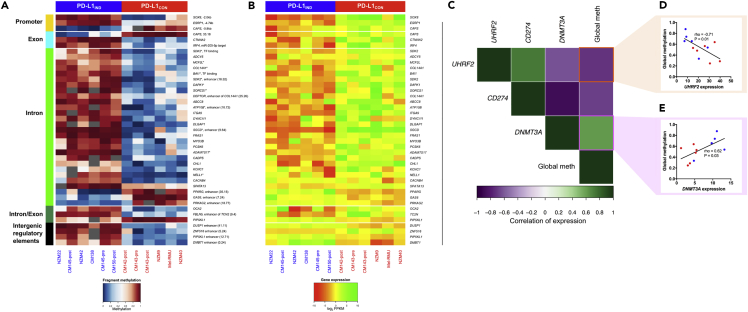
Figure 4.
Differential Methylation Pattern and Relationship with Differential Expression in PD-L1IND and PD-L1CON Cell Lines and Role of Epigenetic Regulators
(A) Methylation heatmap of PD-L1CON and PD-L1IND cell lines for the differentially methylated fragments (DMFs) in different genomic elements showing the relationship with differential mRNA expression (blue = unmethylated, red = fully methylated). For several genes, multiple DMFs showed a strong correlation and are indicated as *.
(B) Mean-centered heatmap of the expression level (log2 FPKMs) of the 39 genes that are regulated by methylation levels.
(C) Correlogram showing cross-correlation of the key epigenetic regulator genes with CD274 (PD-L1) expression and global RRBS methylome levels.
(D and E) Relationship between mRNA level and RRBS methylome for the analyzed cell lines for de novo methylation machinery genes UHRF2 (D) and DNMT3A (E). Spearman rho and statistical significance are shown.
See also Figures S13–S15 and Table S5.