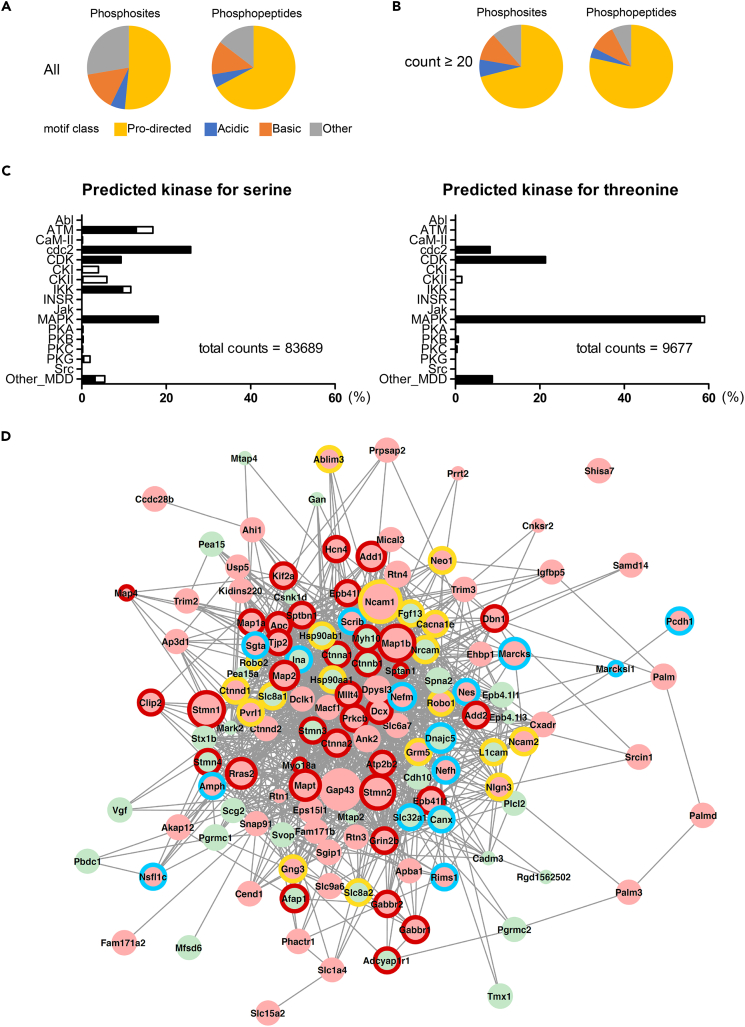
Figure 2.
Bioinformatics Analysis of Phosphosites Identified by GCM Phosphoproteomics of Rat P1 Brain Reveals that P-Directed Sites Are Mainly Dependent on MAPK
(A and B) Fractions of phosphosites (left) and phosphopeptides (right) that are substrates of acid, basic, and proline-directed kinases for all data (A) or data thresholded by 20 counts (B).
(C) Protein kinases predicted for the serine (left) and threonine (right) phosphosites using KinasePhos server. The value in each row represents the fraction of phosphopeptides that are the targets of each kinase. The fractions were further divided into P-directed (filled bars) or non-P-directed (open bars) phosphorylation.
(D) Protein association network for P-directed and non-P-directed proteins. Protein association network was constructed using the STRING database (Szklarczyk et al., 2017), merged with data from human, rat, and mouse. Red and green filled circles indicate P-directed and non-P-directed phosphorylated proteins, respectively. The size of the circle for each protein represents its phosphorylation frequency in GCM. The colors of the external rings indicate enriched protein network groups: group I (red), cytoskeletal proteins (microtubule-related proteins, cortical skeletal proteins, and actin-binding proteins); group II (yellow), signaling molecules related to axon growth/guidance (cell adhesion molecules, proteins in cAMP- or Ca2+-dependent signaling pathways, small GTPase signaling molecules, and guidance receptors); and group III (blue), other categories. Proteins without the external rings were not enriched.