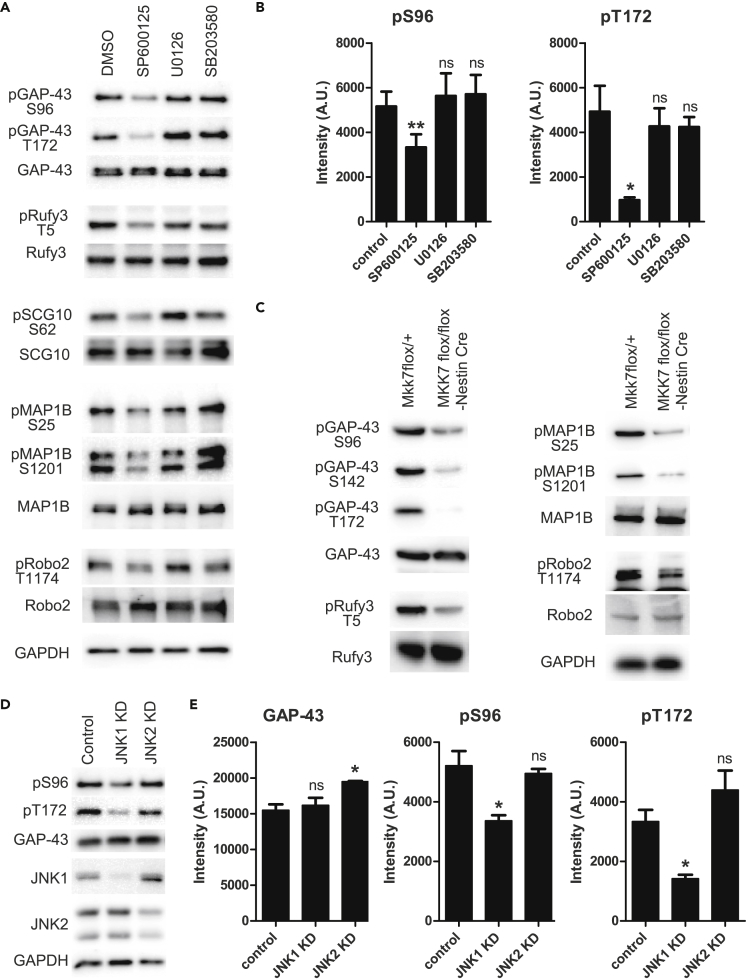
Figure 3.
MAPK Substrates Identified by GCM Phosphoproteomics Undergo JNK-Dependent Phosphorylation
(A and B) Mouse cortical neurons were treated with 20 μM SP600125 (JNK inhibitor), 5 μM U0126 (MEK1/2 inhibitor), or 5 μM SB203580 (p38 inhibitor) for 3 hr. As a control, an equal volume of the solvent DMSO was added to the medium. (A) The SP/TP phosphorylated sites of various GCM proteins are JNK dependent. Frequencies not appearing in Table S1 are as follows: Robo2 [pT1154] = 25; GAP-43 [pS142] = 18; and Rufy3 [pT5] = 19. Western blot results of non-phosphospecific Abs are shown as negative controls. Kinase inhibitors did not affect the reactivity of any of these non-phosphospecific Abs. (B) Effects of MAPK inhibitors on GAP-43 phosphorylation at S96 and T172. Values represent the measured intensity (mean ± SEM, n = 3). **p < 0.01; *p < 0.05; ns, p > 0.05. One-way repeated measures ANOVA with Bonferroni tests to the control.
(C) Brain-specific cKO of MKK7 (Yamasaki et al., 2011), an upstream activator of JNK, suppressed the identified SP/TP phosphorylation. Brain extracts from WT and MKK7flox/flox Nestin-Cre embryos at E15.5 were analyzed by immunoblotting using the indicated Abs. Western blot results of non-phosphospecific Abs are shown as the controls. Kinase inhibitors did not affect the reactivity of any of these non-phosphospecific Abs.
(D and E) Effects of mouse JNK knock down on GAP-43 pS96 and pT172. The representative western blotting (D) and quantified (E) data. Values represent the measured intensity (mean ± SEM, n = 3). *p < 0.05; ns, p > 0.05. One-way ANOVA with Bonferroni tests to the control. GAPDH: glyceroaldehyde-3-phosphate dehydrogenase (A, C, and D).