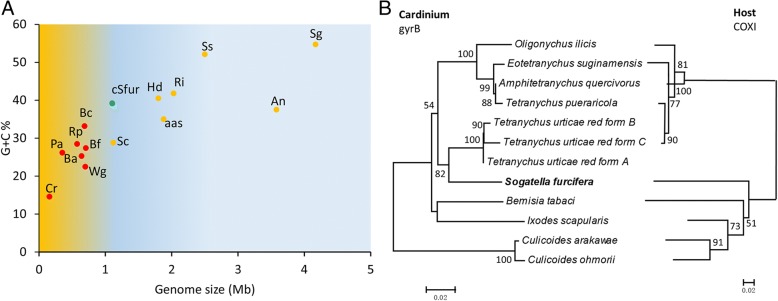
Fig. 8.
Genome size comparison and evaluation of evolutionary relationships between Cardinium and their hosts. a Comparison of genome sizes and GC content of selected endosymbiotic bacteria. P-endosymbionts are marked in red, S-endosymbionts in orange, and Cardinium cSfur is specially marked in green. Cr, Carsonella ruddii DC; Pa, Portiera aleyrodidarum BT-B-HRs; Rp, Riesia pediculicola; Ba, Buchnera aphidicola str. Sg; Bc, Baumannia cicadellinicola; Bf, Blochmannia floridanus; Wg, Wigglesworthia glossinidia; Sc, Spiroplasma chrysopicola; Hd, Hamiltonella defensa 5AT; aas, Amoebophilus asiaticus 5a2; Ri, Regiella insecticola R5.15; Ss, Serratia symbiotica str. Tucson; An, Arsenophonus nasoniae; Sg, Sodalis glossinidius. b Comparisons of the evolutionary relationships between Cardinium strains and their different insect hosts. The host tree is based on the mitochondria gene COXI sequence, while the endosymbiont Cardinium tree is based on gyrB