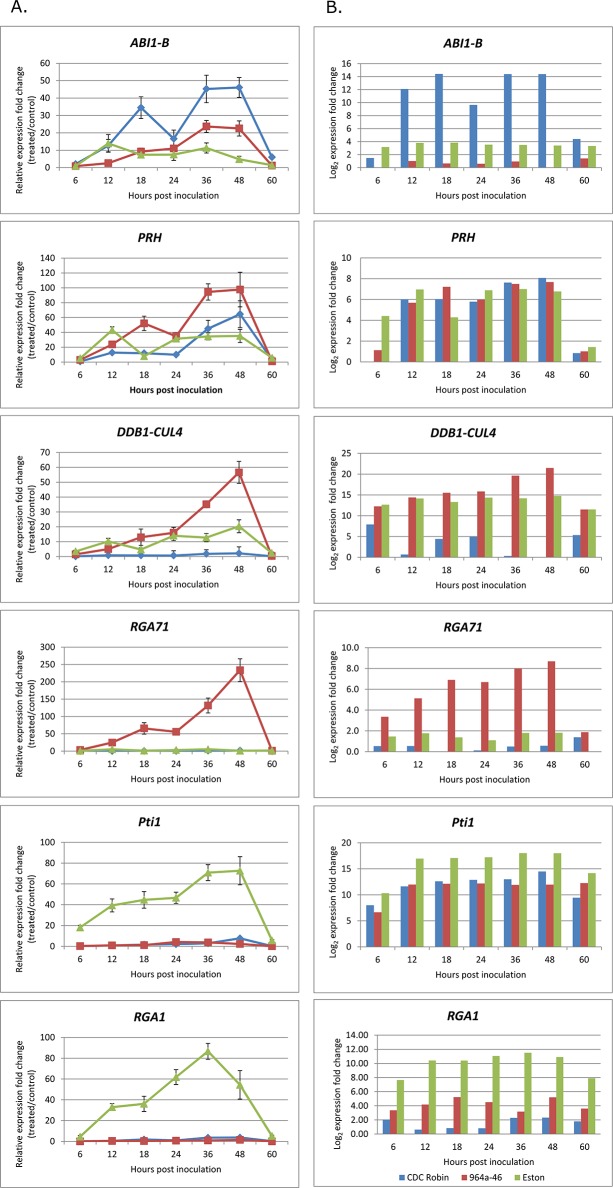
Fig 5. Differential expression of Abscisic acid insensitive 1b (ABI1-B), ddb1- and cul4-associated factor (DDB1-CUl4), pathogenesis-related homeodomain (PRH), Pseudomonas syringae pv. tomato R-gene (Pto)-interactor 1 (Pti1), resistance gene analogue 1 (RGA1) and resistance gene analogues 71 (RGA71) among lentil genotypes infected with Ascochyta lentis.
A. Expression data (means and standard deviations) measured by quantitative real-time PCR (qRT-PCR) based on three biological replicates. Cycle of threshold (Ct) values were normalized with plant β-actin gene values as a reference gene and the gene expression was reported relative to non-infected plants sampled before inoculation (mock). B. RNA-seq data were generated from samples of one of the three biological replicates used for qRT-PCR. The log2 fold change in gene expression was calculated with Cuffdiff software by dividing fragments per kb of exon per million mapped reads (FPKM) value of infected samples to that of mock.