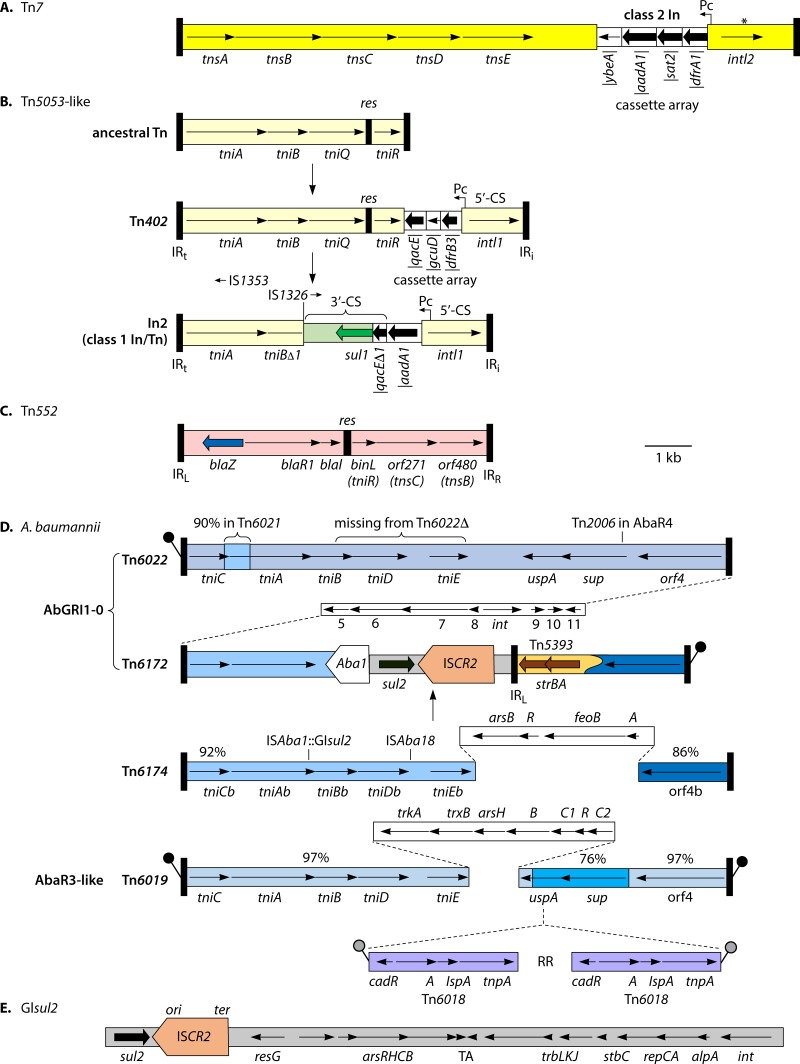
FIG 4.
Tn7-like transposons. Most features are shown as described in the legend to Fig. 3. (A) Tn7. The asterisk indicates the position of a common stop codon in intI2. The diagram was drawn based on the sequence from INSDC accession number AP002527. (B) Evolution of class 1 In/Tn. The diagrams show capture of intI1/attI1/Pc and gene cassettes, with qacE in the last position, by a Tn5053-like transposon. Subsequent deletion of parts of the final qacE cassette and tni region and insertion of sul1 create the 3′-CS, giving a typical “clinical” class 1 In/Tn which is not self-transposable, e.g., In2. Diagrams are based on information in reference 21 and sequences from INSDC accession numbers U67194 and AF071413. Different extents of tni and different IS may be present beyond the 3′-CS (see Fig. 5 in reference 21 for further details). (C) Tn552. Gene names shown in parentheses indicate relationships to those in Tn7/Tn5053 elements. The diagram was drawn based on the sequence from INSDC accession number X52734. (D) Transposons making up resistance islands in A. baumannii. The top diagram represents Tn6022; differences in minor variants Tn6021 (a short region with only 90% identity matches Tn6172) and Tn6022Δ are shown. The main part of the Tn6174 diagram corresponds to the hypothetical, ancestral Tn6173, which is also related to Tn6022 (percentage identities in different regions shown above) but has the ars/feo region replacing uspA and sup. Tn6174 itself has the two insertions shown above the diagram. Tn6172 was generated from Tn6174 by addition of Tn5393 (Fig. 3) and an internal deletion. In AbGRI1-0, a region flanked by Tn6022 and Tn6172 is inserted into the chromosomal comM gene. The backbone Tn6019 of AbaR3-like islands is related to Tn6022 (percent identities in different regions are shown) but contains an additional segment, shown above the relevant diagram. Various regions containing different antibiotic resistance genes are found between the two copies of Tn6018 (designated RR). Diagrams are based on previously published information (109, 110) and on sequences from the following INSDC accession numbers: Tn6022, CP012952; Tn6021 and Tn6164, CP012005; Tn6022Δ, JN247441; Tn6172, KU744946; and Tn6019, FJ172370. (E) GIsul2 (15.460 kb [188] rather than the initially reported 15.456 kb [61], apparently due to errors in the S. flexneri sequence). ars, arenite/arsenate resistance gene; TA, toxin-antitoxin system; alpA, regulation gene. The diagram is based on information in reference 188 and the sequence from INSDC accession number KX709966. The resistance genes shown confer resistance to the following antibiotics: aadA1, streptomycin and spectinomycin; sat2, streptothricin; dfrA1 and dfrB3, trimethoprim; qacE, quaternary ammonium compounds; sul1 and sul2, sulfonamides; blaZ, penicillins; and strAB, streptomycin.