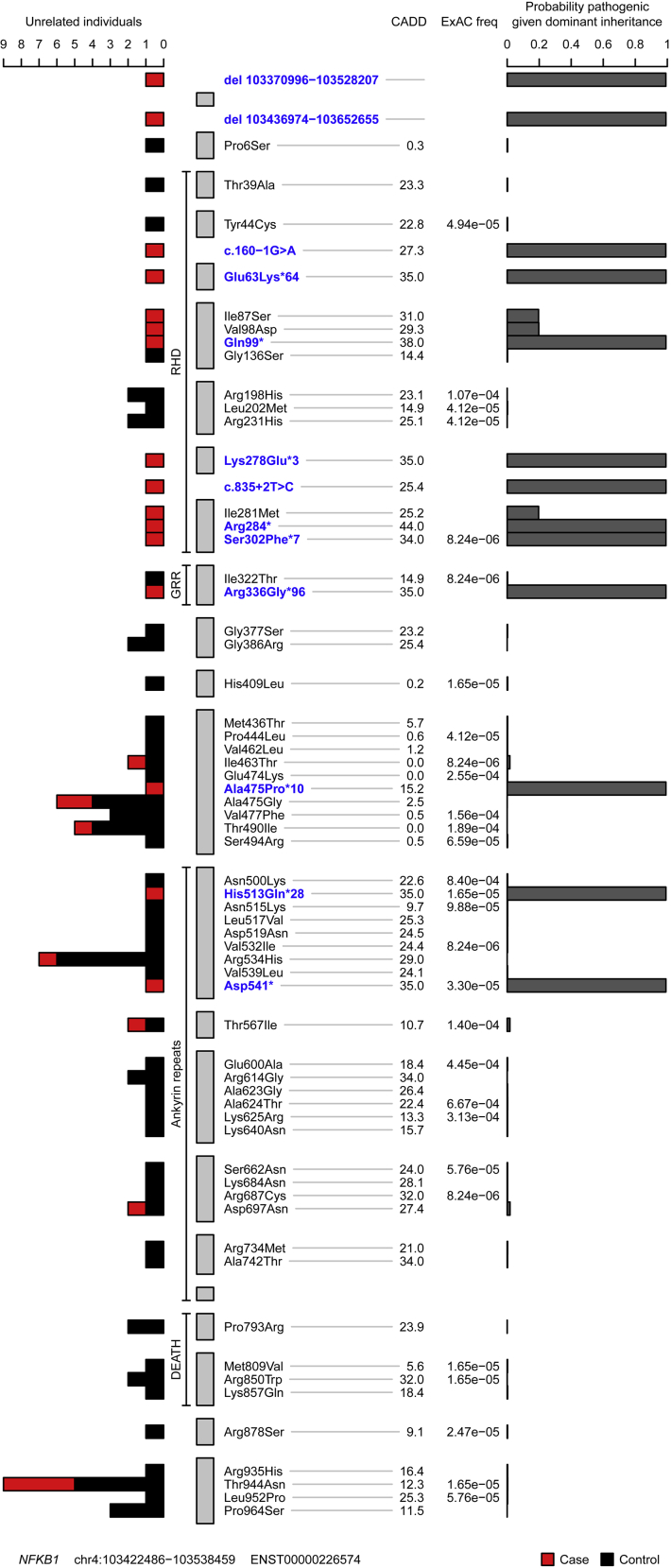
Fig 2.
Plot of rare missense, truncating, and gene deletion NFKB1 variants identified in the NIHRBR-RD genomes of unrelated subjects and their location relative to NFKB1 domains. Tracks from left to right show the following: number of unrelated case (red) and control (black) subjects in whom each variant was observed; the 4 major NFKB1 domains; each exon in transcript ENST00000226574 (gray bars); variant annotation relative to transcript ENST00000226574 and genomic location of large deletions, with VEP high-effect variants and large deletions highlighted in blue; Combined Annotation Dependent Depletion (CADD) scores of all nonsense, frameshift, splice and missense variants; Exome Aggregation Consortium (ExAC) allele frequencies; and conditional probability of variant pathogenicity inferred by using BeviMed. Only variants labeled as being of moderate or high effect relative to the canonical transcript ENST00000226574 are shown. The initial inference that formed part of the genome-wide analysis included variant chr4:103423325G>A, which was observed in 1 control sample. This variant is intronic (low effect) relative to ENST00000226574 but is a splice variant (high effect) relative to the minor transcript ENST00000505458. Because variants were filtered based on the highest-effect variant annotation against any Ensembl transcript, this variant was originally included in the inference. For this plot, the inference was rerun, including only missense, truncating, and gene deletion variants relative to the canonical transcript.