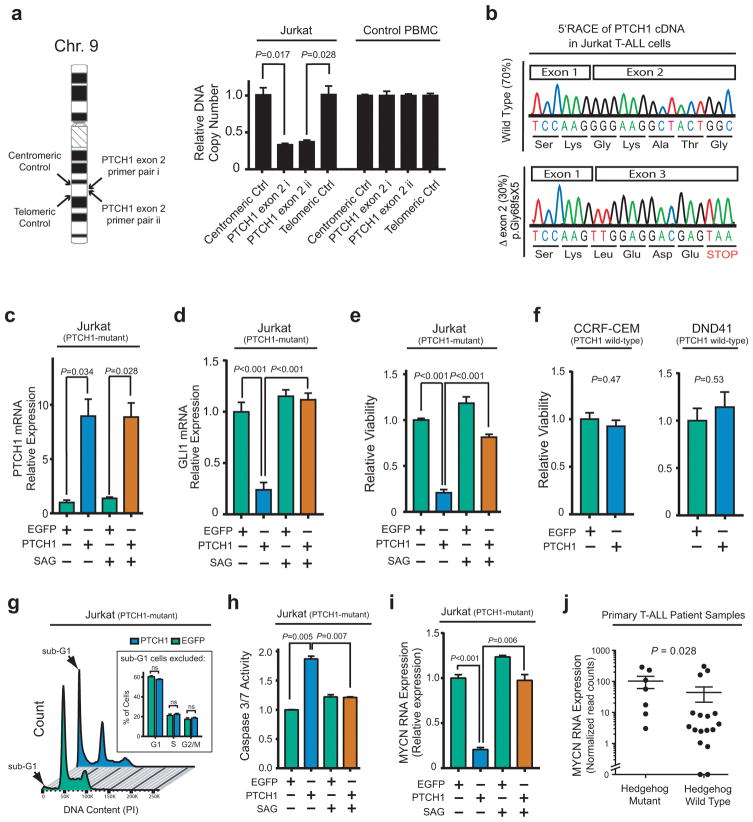
Figure 2. Aberrant hedgehog pathway activation is required for survival of PTCH1-mutant T-ALL cells.
(a) Quantitative DNA PCR analysis of DNA copy number using independent primer pairs located within exon 2 of PTCH1 (NM_000264), as well as centromeric and telomeric primer pair controls (location shown on left), on genomic DNA from Jurkat T-ALL cells. (b) 5′RACE analysis of PTCH1 mRNA transcripts expressed in Jurkat T-ALL cells. (c–d) Jurkat cells were transduced with wild-type PTCH1 or EGFP control, and treated with DMSO control or 100nM SAG to activate the Hedgehog pathway downstream of PTCH1. Quantitative reverse transcriptase PCR analysis was performed to assess mRNA expression of PTCH1 (c) or GLI1 (d), a canonical reporter of Hedgehog pathway activity. Results were normalized to β-Actin. A two-sided Welch t-test was used for statistical analysis of PTCH1 mRNA overexpression (c), and an ANOVA with Tukey adjustment for multiple comparisons was performed to assess differences in GLI1 mRNA expression (d). (e–f) Viability of the indicated T-ALL cells transduced with EGFP control or wild-type PTCH1, assessed 48 hours after selection in puromycin using trypan blue staining. A two-sided Welch t-test was used for statistical analysis. A Bonferroni correction was applied to adjust for multiple hypotheses testing (e). (g) Jurkat cells transduced with PTCH1 or EGFP control were harvested 48 hours after selection in puromycin and stained with PI for cell cycle analysis. A two-way ANOVA with Tukey adjustment was applied to assess differences in G1, S, and G2/M populations; p-values for each comparison in these three populations were not significant. (h) Caspase 3/7 activity in Jurkat cells transduced with PTCH1 versus EGFP control, and treated with 100 nM SAG or vehicle control (DMSO); results were normalized to DMSO-treated EGFP control. A two-sided Welch t-test with Bonferroni correction was applied for statistical analysis. (i) Relative MYCN mRNA expression (normalized to β-Actin control) was assessed by RT-PCR in Jurkat cells transduced with either EGFP or wild-type PTCH1, and treated with DMSO control or 100nM SAG to activate the Hedgehog pathway downstream of PTCH1. A two-sided Welch t-test with Bonferroni correction was used to assess significance. (j) Association of MYCN mRNA expression assessed using RNA sequencing analysis in 24 of the 109 patients in this study, with Hedgehog pathway mutations. A Wilcoxon sum-rank test was used to assess significance.