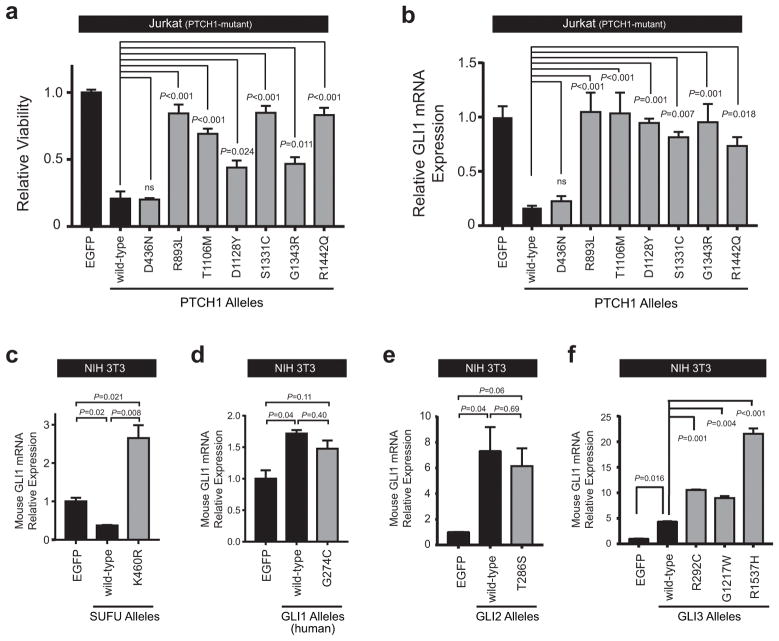
Figure 3. Most Hedgehog pathway mutations encode pathogenic alleles.
(a) Jurkat cells were transduced with EGFP control, wild-type PTCH1, or PTCH1 mutant alleles identified within our patient cohort, and viability was assessed following selection in puromycin on day 5 using trypan blue staining. Results were normalized to EGFP control. A two-sided, one-way ANOVA with Dunnett’s adjustment for multiple comparisons was used for statistical analysis. (b) Relative GLI1 mRNA expression (normalized to β-Actin control) was assessed in Jurkat cells transduced with EGFP, wild-type PTCH1, or mutant PTCH1 alleles. A one-way ANOVA with Dunnett’s adjustment for multiple comparisons was used for statistical analysis. NIH 3T3 mouse fibroblast cells were transfected with EGFP control, (c) wild-type SUFU or K460R mutant SUFU allele, (d) wild-type GLI1 or G274C mutant GLI1 allele, (e) wild-type GLI2 or the T286S mutant GLI2 allele, or (f) wild-type GLI3, or one of GLI3 mutant alleles (R292C, G1217W, R1537H) identified within our T-ALL patient cohort, and relative mRNA expression of mouse Gli1 (normalized to mouse Gapdh control) was assessed using RT-PCR. Results were normalized to EGFP control. A one-way ANOVA with Dunnett’s adjustment for multiple comparisons was used for statistical analysis.