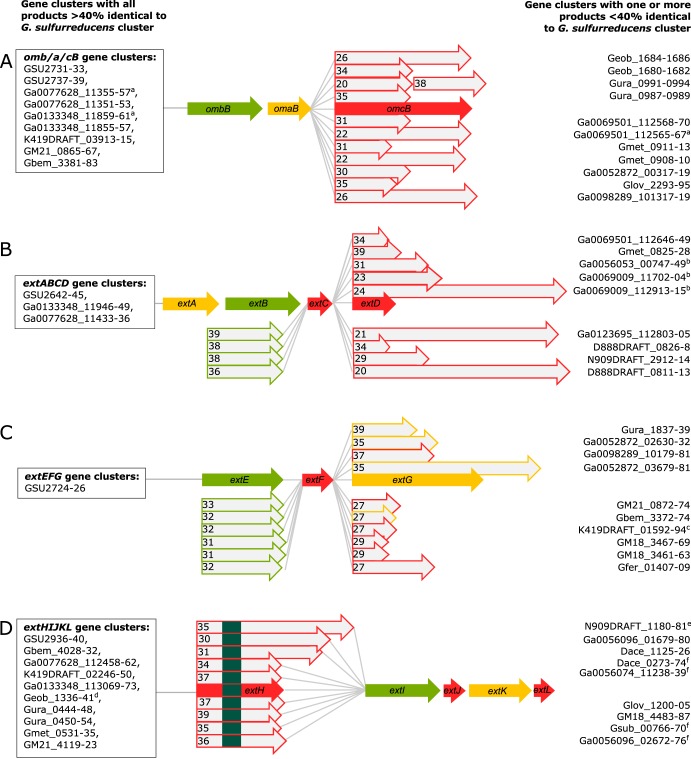
FIG 9.
Cytochrome conduit conservation across the order Desulfuromonodales. Shown is a representation of cytochrome conduit clusters from the Desulfuromonodales with homologs to either OmcBC (A), ExtABCD (B), ExtEFG (C), or ExtHIJKL (D). Complete clusters with all components sharing >40% identity to the corresponding G. sulfurreducens cytochrome conduit are indicated in boxes to the left of each gene cluster. Clusters in which one or more proteins are replaced by a new element with <40% identity are listed on the right side of each gene cluster. Numbers with proteins indicate the percent identity to the G. sulfurreducens version. Red arrows, putative outer membrane products with a predicted lipid attachment site; yellow arrows, predicted periplasmic components; green arrows, predicted outer membrane anchor components. Superscript letters a to d indicate the following: a, OmcBC homologs in these gene clusters also encode Hox hydrogenase complexes; b, gene clusters have contiguous extBCD loci but extA is not in the vicinity, as extA was found in separate parts of the genome for some of those organisms (see Table S2 in the supplemental material); c, the gene cluster has additional lipoprotein decaheme c-type cytochrome upstream of extE; d, lipid attachment sites corresponding to ExtJL could not be found, but there is an additional small lipoprotein encoded within the gene cluster. For ExtHIJKL clusters, homologs depicted above extH are found in gene clusters containing only extI, whereas homologs depicted below extH are found in gene clusters containing full extHIJKL loci. Upstream and on the opposite strand to all gene clusters homologous to extHIJKL there is a transcription regulator of the LysR family, except where marked by superscript letter e, where there is no transcriptional regulator in that region, and superscript letter f, where there are transcriptional regulators of the TetR family instead.