Summary
Ovarian cancers are known to evade immunosurveillance and to orchestrate a suppressive immune microenvironment. Here we examine the role of human epididymis protein 4 (HE4), an ovarian cancer biomarker, in immune evasion. Through modified subtractive hybridization analyses we have characterized the gene targets of HE4 in human peripheral blood mononuclear cells (PBMCs), and established a preliminary mechanism for HE4‐mediated immune failure in ovarian tumours. Upon exposure of purified PMBCs to HE4, osteopontin (OPN) and dual‐specificity phosphatase 6 (DUSP6) emerged as the most suppressed and up‐regulated genes, respectively. SKOV3 and OVCAR8, human ovarian carcinoma cell lines, exhibited enhanced proliferation in conditioned media from HE4‐exposed PBMCs, an effect that was attenuated by the addition of recombinant OPN or OPN‐inducible cytokines [interleukin (IL)‐12 and interferon (IFN)‐Ɣ]. Additionally, upon co‐culture with PBMCs, HE4‐silenced SKOV3 cells were found to be more susceptible to cytotoxic cell death. The relationship between HE4 and OPN was reinforced further through the analysis of serous ovarian cancer patient samples. In these biopsy specimens, the number of OPN+ T cells correlated positively with progression free survival (PFS) and inversely with serum HE4 level. Taken together, these findings show that HE4 enhances ovarian cancer tumorigenesis by compromising OPN‐mediated T cell activation.
Keywords: cancer, cytokines, T cells
Introduction
Human epididymis protein 4 (HE4) is a family member of the whey acidic proteins (WAPs), which share a four‐disulphide core domain and are generally regarded as protease inhibitors 1, 2, 3. HE4 was first identified in the male reproductive tract but has since been found in select other tissues, such as the kidney, female reproductive tract, breast and lungs 4, 5. In addition, it is highly over‐expressed in several human malignancies, including ovarian and endometrial cancers 5, 6, 7, 8. HE4’s exact role in normal and malignant tissue is still unclear; however, as a negative prognostic factor in women with epithelial ovarian cancer, its serum levels correlate with chemoresistance and reduced overall survival 9, 10, 11. Our previous work with HE4 has led to the development of a United States Food and Drug Administration (US FDA)‐approved biomarker tool for the evaluation of pelvic masses, named the ‘risk of ovarian malignancy algorithm’ or ROMA 12, 13, 14, 15. The ROMA score incorporates HE4, cancer antigen 125 (CA‐125) and menopausal status into a calculation to estimate ovarian cancer risk. As a biomarker, HE4 detection and monitoring is already improving patient care, yet it is imperative that we learn more about its function in order to better understand ovarian tumorigenesis and ultimately develop effective therapies for this highly fatal cancer.
Through this present study, we have begun to elucidate HE4’s role in tumour immune suppression. We have identified differentially expressed genes in peripheral blood mononuclear cells (PBMCs) after exposure to HE4 using a modified subtractive hybridization method. This strategy identified osteopontin (OPN) as one of the most prominently suppressed targets in PBMCs following HE4 treatment. OPN is a secretory glycosylated phosphoprotein encoded by the gene SPP1. OPN contains an arginine–glycine–aspartate (RGD) sequence that, via interactions with integrin family members or CD44, triggers downstream signalling events and relays early cell‐mediated immune responses 16, 17, 18. We have observed that HE4‐induced OPN suppression diminishes the cytotoxicity of PBMCs against cultured human ovarian cancer cells in vitro. Further, the expression levels of OPN in stromal infiltrating T cells in biopsy samples from serous ovarian cancer patients show a direct association with patient progression‐free survival (PFS). Taken together, our data here demonstrate that HE4 can inhibit the anti‐tumoral immune function of PBMCs, most prominently T cells, via suppression of OPN production.
Materials and methods
Subtractive hybridization and thymine and adenine (TA) cloning
Primary human PBMCs were obtained from a single healthy volunteer donor under Institutional Review Board (IRB) approval from the Women and Infants Hospital. Approximately 5 × 107 PBMCs were isolated from 40 ml of heparinized total blood, resuspended in 5 ml of serum‐free RPMI‐1640 medium (no. 31800022; Invitrogen, Carlsbad, CA, USA) and incubated with 0·01 μg/ml, approximately 270 pm, of rHE4 (MBS717359; MyBiosource, San Diego, CA, USA) or diluent control for 6 h. Total RNA was isolated using TRIzol reagent (Invitrogen). HE4 was found to have dose‐dependent actions on PBMCs (Supporting information, Fig. S1a,b), so we chose a physiologically relevant concentration of 270 pm, which correlates with ovarian cancer patient serum HE4 levels 19; 300 μg of total RNA was isolated and then stored at –80°C. Blood collection and HE4 exposure was repeated every 7+ days until a total of 1 mg RNA was collected from each treatment group. Approximately 10 μg of mRNA was purified from the total RNA using oligo‐dT‐coated magnetic beads (Takara‐Clontech, Mountain View, CA, USA). Five μg mRNA was used to construct subtractive cDNA libraries using the polymerase chain reaction (PCR)‐SelectTM cDNA Subtraction Kit (Takara‐Clontech), following the manufacturer’s instructions. Briefly, the tester and driver cDNAs were synthesized from poly A+ RNA generated from control and HE4‐treated PBMCs. The tester and driver cDNAs were digested with restriction enzyme, Rsa I, to yield shorter, blunt‐ended molecules. The tester cDNA was then subdivided into two portions, and each was ligated to a different cDNA adaptor. The ends of the adaptors do not contain phosphate groups, so only one strand of each adaptor attaches to the 5' end of the cDNA. The two adaptors have stretches of identical sequence to allow annealing of the PCR primer once the recessed ends have been filled in. The differentially expressed genes were identified through two steps of hybridization and PCR. The detailed procedure is described in Supporting information, Fig. S2a. The PCR products of the differentially expressed genes were cloned into a pUC19‐TA vector, selected by blue/white selection, amplified by colony PCR using M13 primers (Supporting information, Table S2), and subjected to direct sequencing for PCR products in the range of 200–3000 base pairs (Supporting information, Fig. S2b).
Cell culture
The human ovarian tumour cell lines, SKOV3 and OVCAR8, were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). RPMI‐1640 (no. 31800022; Invitrogen) and Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 1·0 mM sodium pyruvate (no. 31600034; Invitrogen) were used for culturing PBMC and tumour cells, respectively. Conditioned media were obtained from a 24‐h PBMC exposure to 0·01 μg/ml (270 pm) of rHE4 versus control. Residual rHE4 in the conditioned media was depleted as follows: 5 ml of media was incubated with 10 μg (100 μl) of anti‐human HE4 antibody (sc‐293473; Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h at 4°C, 100 μλ packed volume of protein G‐coated sepharose beads (GE Healthcare Life Science, Pittsburgh, PA, USA) were added and incubated for an additional 4 h. After incubation, the sepharose beads were removed by centrifugation and the supernatant was processed through a sterile 0·2 μm pore syringe filter. The depletion of HE4 from the conditioned media was confirmed by enzyme‐linked immunosorbent assay (ELISA) (Supporting information, Table S3). For the cell‐mediated cytotoxicity assay, 2 × 105 target cells (scrambled control plasmid or shHE4 transfected SKOV3) were seeded in six‐well plates and incubated overnight with complete media. The next day, cells were placed in serum‐free media for another overnight incubation to induce quiescence before 1 × 107 PBMCs, mixed with propidium iodide (Invitrogen), were added to each well. Some of the wells contained 5 pg/ml of recombinant IL‐12 (rIL‐12) (219‐IL‐005; R&D Systems, Minneapolis, MN, USA), 20 pg/ml of recombinant IFN (rIFN)‐Ɣ (SRP3058; Sigma‐Aldrich, St Louis, MO, USA) or 0·05 μg/ml of rHE4 (ab132299; Abcam, Cambridge, UK) in combination, as indicated in Fig. 4 (lower panel). In order to avoid confounding additives from serum, all experiments were performed under serum‐free conditions. The ovarian cancer cell lines appeared viable and morphologically unchanged for up to 72 h in serum‐deprived DMEM. Short hairpin RNA (shRNA) for human HE4 (TR318721; Origene, Minneapolis, MN, USA) was transfected into SKOV3 using lipofectamine 2000TM (Invitrogen), following the manufacturer’s instructions. Also, where indicated cells were treated with 20 pg/ml of recombinant OPN (ab92964; Abcam) or 0·01 μg/ml rHE4.
Figure 4.
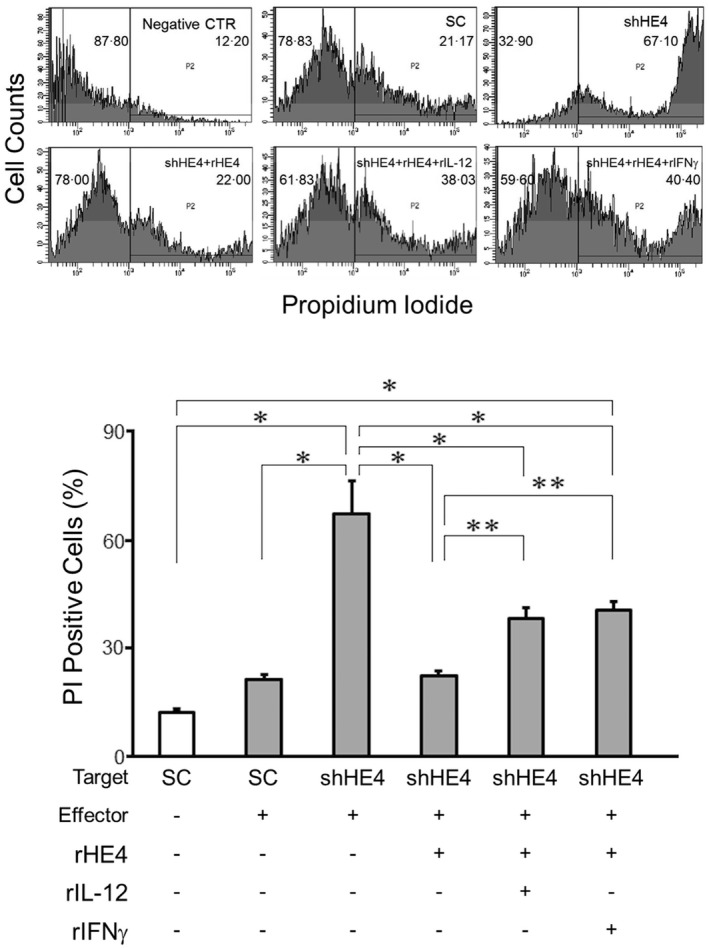
Flow cytometric analysis of the cytotoxicity of mononuclear cells against SKOV3 tumour cells. Cell membranes of SKOV3 (target) cells were labelled with DiOC18(3) fluorescent dye and then incubated with peripheral blood mononuclear cells (PBMCs) in the presence of propidium iodide (PI) as a marker of cell death. After washing away the non‐adherent cells (PBMCs), the PI‐positive tumour cells were quantitated via flow cytometry (upper panel). The numbers on the histograms represent mean percentage of each bisection. The bar graph (lower panel) represents percentages of PI‐positive (dead/dying) cells in various culture conditions. The mean is shown; error bars represent standard error of the mean (s.e.m.) (n > 10). *P < 0·01; **P < 0·05.
Quantitative real‐time PCR (qRT–PCR)
RNA was isolated from the PBMCs of healthy donors using TRIzol (Invitrogen), according to the manufacturer’s instructions. cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen). qPCR was performed using Premix Ex‐TaqTM II (Clontech‐Takara, Mountain View, CA, USA) probes for OPN, IL‐12β and IFN‐Ɣ. All reactions were normalized using glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) as an endogenous control. Amplification data were analysed using the ΔΔ Ct method. Sequences of PCR primers are summarized in Supporting information, Table S2.
Flow cytometry
Fluorescein isothiocyanate (FITC)‐labelled anti‐CD3 (HIT3a), CD14 (M5E2), CD19 (HIB19) and CD56 (B159) antibodies were obtained from BD Biosciences (Billerica, MA, USA). Alexa Fluor® 647‐labelled anti OPN antibody (EPR3688) was obtained from Abcam. After staining for cell surface markers (CD3, CD14, CD19 and CD56) the cell membrane was permeabilized by 0·2% Triton X‐100 and 0·2% digitonin, and then stained for OPN. Flow cytometric analysis was performed with the fluorescence activated cell sorter (FACS)Canto system, utilizing FACSDiva software (BD Biosciences).
ELISA
ELISA kits for OPN, IL‐12αβ, IFN‐Ɣ and HE4 were obtained from R&D Systems. The assays were performed following the manufacturer’s instructions.
Viability and migration assays
SKOV3 cells, 1 × 103 per well, were seeded in a 96‐well culture plate. After overnight incubation with serum‐free medium, conditioned media was added to the quiescent cells. Cell viability was assessed at 24, 48 and 72 h using CellTiter‐Blue® (Promega, Madison, WI, USA). Cell migration was assessed using the InnoCyteTM Cell Migration Assay (EMD Millipore, Taunton, MA, USA). In short, 5 × 104 SKOV3 cells were seeded in the upper chamber of a 96‐well plate, with the lower chamber containing the PBMC‐conditioned media. Transmigration was determined after incubating the cells for 24 h in a CO2 incubator at 37°C.
Immunohistochemistry
SKOV3 cells, 0·5 × 104 per chamber, were seeded in a four‐chamber slide. After overnight incubation with serum‐free medium, conditioned media was added to the quiescent cells for an additional 48 h. The cells were then fixed and permeabilized with 2% formaldehyde and 0·2% Triton X‐100 and stained with a mouse monoclonal anti‐Ki67 antibody (clone B56; PD Pharmingen, Franklin Lakes, NJ, USA) overnight at 4°C in a humidified chamber. The slides were then incubated with an alkaline phosphatase (ALP)‐conjugated anti‐mouse immunoglobulin (Ig)G (H + L) secondary antibody (Bio‐Rad, Hercules, CA, USA) for 30 min at room temperature. Bound antibody was detected using the ALP substrate kit (Vector Laboratories, Burlingame, CA, USA) and counterstained lightly with veronal acetate‐buffered 1% methyl green solution, pH 4.0 (Vector Laboratories). PermountTM (Fisher Scientific, Ottawa, Ontario, Canada) was used as the mounting media and sections were coverslipped. Immunohistochemical studies were repeated four times on samples prepared from different cultures. The proportion of Ki67‐positive cells was calculated according to the following formula: 100 × (Ki67‐positive nuclei/total nuclei). Each image was analysed at least four times to obtain an average labelling index.
Western blotting
HE4 shRNA transfected SKOV3 cells were lysed and Western blot was performed as described previously 20. Anti‐HE4 antibody was obtained from Origene (TA326648) and anti‐actin antibody (clone 2G2; EMD Millipore) was used for detection of the internal loading control. The results were visualized with SuperSignal™ substrate (ThermoFisher Scientific) and analysed with the UN‐SCAN‐IT gel software for Windows (version 6.1; Silk Scientific, Inc., Orem, UT, USA).
Confocal immunofluorescent microscopy
Formalin‐fixed, paraffin‐embedded tissue sections were cut to a thickness of 4 μm. For heat‐induced epitope retrieval, deparaffinized sections in 0·01 M citrate buffer were treated three times at 90°C for 5 min using a microwave oven. After blocking with 10% normal horse serum, sections were incubated with rabbit anti‐OPN antibody (FL‐314; Santa Cruz Biotechnology) or mouse anti‐CD3 (PS‐1; Abcam) overnight at 4°C. They were then washed with PBS and incubated with DyLight 488 goat anti‐rabbit IgG (DL1488; Vector Laboratories) or DyLight 594 horse anti‐mouse IgG (DL2594; Vector Laboratories) secondary antibodies for 1 h at room temperature in the dark. Slides were washed again with PBS and coverslipped with DAPI‐containing mounting medium (Vector Laboratories). Confocal images were acquired with a Nikon C1si confocal (Nikon Inc., Mellville, NY, USA) using diode lasers 402, 488 and 561. Ten fields/sample were selected randomly based on DAPI staining and counts were performed for CD3 and OPN using a ×40 objective. Counts are expressed as the number of positive cells/mm2.
All human tissue and blood donors provided written informed consent. The study was approved by the Women and Infants Hospital ethics committee.
Statistics
Data were expressed as an average ± standard error of the mean (s.e.m.) of at least three independent experiments. An unpaired, two‐tailed Student’s t‐test was used to determine significance. Multiple treatments were analysed by using one‐way analysis of variance (anova) followed by Ryan’s multiple comparison test. Spearman’s rank correlation test was used to assess the immunofluorescent staining on biopsy specimens. Differences between groups were considered statistically significant when P < 0·05.
Results
Differential gene expression in PBMCs after HE4 exposure
To identify HE4’s impact on gene expression in PBMCs, modified subtractive hybridization was performed as depicted in Supporting information, Fig. S2a. PCR products of the differentially expressed genes were cloned into pUC19‐TA vectors to create a differential cDNA library (Supporting information, Fig. S2b). PCR products from HE4‐induced and suppressed gene colonies were sequenced, resulting ultimately in the identification of 211 up‐regulated and 208 down‐regulated genes. Among the identified genes, 23 induced and 15 suppressed sequences showed no significant similarity (NSS) to known genes in available nucleotide databases. The differentially expressed genes were classified by their function as summarized in Supporting information, Table S1. Among the 208 suppressed genes, OPN emerged as the most frequently identified gene (six times out of 253 sequences, 2·4%; Table 1).
Table 1.
Genes suppressed in response to human epididymis protein 4 (HE4)
| Frequency | ID | Gene name |
|---|---|---|
| 15 | NSS | No significant similarity |
| 6 | NM_001040058 | Secreted phosphoprotein 1 (SPP1), transcript variant 1 |
| 3 | NM_015574 | Ankyrin repeat domain 17 (ANKRD17) |
| 3 | NM_001693 | ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 (ATP6V1B2) |
| 3 | NM_000206 | Interleukin 2 receptor subunit gamma (IL2RG) |
| 3 | NM_022818 | Microtubule‐associated protein 1 light chain 3 beta (MAP1LC3B) |
| 3 | NM_001243121 | Phosphodiesterase 4A (PDE4A) |
| 3 | NM_080792 | Signal regulatory protein alpha (SIRPA) |
| 3 | NM_015131 | WD repeat domain 43 (WDR43) |
| 2 | NM_001025604 | Arrestin domain containing 2 (ARRDC2) |
| 2 | NM_001164755 | Aspartate beta‐hydroxylase (ASPH) |
| 2 | NM_032408 | Bromodomain adjacent to zinc finger domain, 1B (BAZ1B) for peer review |
| 2 | NM_002985 | Chemokine (C‐C motif) ligand 5 |
| 2 | AC132216 | Chromosome 11, clone RP13‐786C16 |
| 2 | NC_018926 | Chromosome 15, alternate assembly CHM1_1.1 |
| 2 | NM_001291549 | Cyclin‐dependent kinase inhibitor 1A (p21, Cip1) (CDKN1A) |
| 2 | NM_014280 | DnaJ (Hsp40) homologue, subfamily C, member 8 (DNAJC8) |
| 2 | XM_011535514 | Eukaryotic translation elongation factor 1 alpha 1 (EEF1A1) |
| 2 | XM_011518416 | Family with sequence similarity 120A (FAM120A) |
| 2 | NM_020447 | Family with sequence similarity 219 member B (FAM219B) |
| 2 | NG_029887 | Golgin A3 (GOLGA3) |
| 2 | NM_002107 | H3 histone, family 3A (H3F3A) |
| 2 | NM_001128619 | Leucine zipper protein 6 (LUZP6) |
| 2 | NM_002463 | MX dynamin‐like GTPase 2 (MX2) |
| 2 | NM_004687 | Myotubularin‐related protein 4 (MTMR4) |
| 2 | NM_001251855 | Phosphoinositide‐3‐kinase regulatory subunit 5 (PIK3R5) |
| 2 | NM_000437 | Platelet‐activating factor acetylhydrolase 2, 40kDa (PAFAH2), |
| 2 | NR_049751 | Reticulon 3 (RTN3) |
| 2 | NM_001198719 | Retinoblastoma binding protein 7 (RBBP7) |
| 2 | NM_001028 | Ribosomal protein S25 (RPS25) |
| 2 | XM_011534644 | Serine/threonine kinase 10 (STK10) |
| 2 | NM_001242933 | Sorting nexin 1 (SNX1) |
| 2 | XR_241300 | Splicing factor 3b, subunit 1, 155kDa (SF3B1) |
| 2 | NM_181892 | Ubiquitin‐conjugating enzyme E2D 3 (UBE2D3) |
| 2 | NM_006007 | Zinc finger, AN1‐type domain 5 (ZFAND5) |
| 1 | 159 genes |
HE4 reduces OPN expression in PBMCs
HE4‐induced suppression of osteopontin in PBMCs was then confirmed via three modalities: flow cytometry, qPCR and ELISA. PBMCs from four individual human donors were used. First, cells were cultured with recombinant human HE4 for 24 h and collected for flow cytometry analysis. Intracellular protein expression of OPN was found to be reduced significantly upon HE4 exposure in T cells (48·8 ± 1·0% versus 37·4 ± 1·9%, Fig. 1a), but not in monocytes, B cells or NK cells (Supporting information, Fig. S3a,b). Osteopontin gene expression was then determined using qPCR, which showed a 0·7‐fold downregulation (Fig. 1b) upon HE4 exposure. ELISA was used to quantify OPN protein concentration in cell lysate and culture supernatant in the presence of HE4. We determined that the concentration of OPN in cell lysate (159·8 ± 3·1 versus 103·6 ± 3·2 pg/ml) and culture supernatant (53·4 ± 3·1 versus 30·1 ± 3·5 pg/ml) were both decreased significantly by HE4 treatment (Fig. 1b,c).
Figure 1.
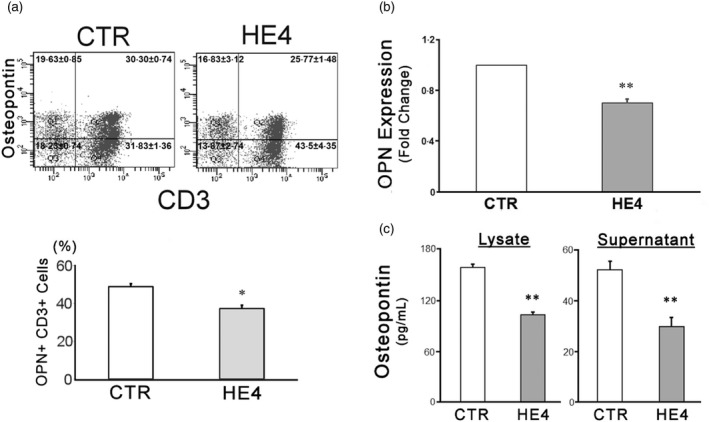
Human epididymis protein 4 (HE4) down‐regulates the expression of osteopontin (OPN) in peripheral blood mononuclear cells (PBMCs). (a) Two‐colour flow cytometric analysis of PBMC following a 24‐h incubation with 0·01 μg/ml of recombinant human epididymis protein 4 (rHE4) or vehicle [control (CTR)]. Two‐dimensional scatterplots of OPN (AlexaFluor 647) and CD3 [fluorescein isothiocyanate (FITC)] are shown. The numbers on the scatterplots represent mean ± standard error of the mean (s.e.m.) % of each quadrant. (b) OPN transcription in response to a 6‐h incubation with 0.01 μg/mL rHE4 (HE4) or vehicle (CTR) were evaluated by real time polymerase chain reaction (PCR). A bar graph represents relative expression levels against control. (c) OPN concentrations of PBMC lysates and culture supernatants after a 24‐h incubation with 0·01 μg/ml of rHE4 or vehicle (CTR) were evaluated by enzyme‐linked immunosorbent assay (ELISA). All the experiments were performed with PBMCs from four individual donors and repeated three (a), nine (b) and 10 (c) times. The mean is shown in the bar graphs; error bars represent s.e.m. (n > 10). *P < 0·05; **P < 0·01.
HE4‐mediates the down‐regulation of IL‐12 and IFN‐Ɣ production, a process that is reversible with OPN supplementation
In lipopolysaccharide‐stimulated macrophages, OPN has been shown to enhance IL‐12 and suppress IL‐10 production, thereby promoting an overall increase in T helper type 1 (Th1) activity 17, 18. In order to determine the impact of HE4‐mediated OPN reduction on Th1 cytokines in PBMCs, IFN‐Ɣ and IL‐12 were evaluated at both the transcriptional and protein levels. Cells were incubated with either: (a) vehicle control, (b) rHE4 or (c) rHE4 and rOPN for 6 h and cell lysates and/or culture supernatants were taken for qPCR and ELISA. As shown in Fig. 2a, the relative expression of IL‐12β and IFN‐Ɣ mRNA was decreased (61 and 69%, respectively) upon treatment with rhHE4. This suppression was reversed partially by the addition of recombinant OPN (rOPN) to culture conditions. Protein expression, as determined by ELISA, is shown in Fig. 2b. IL‐12 concentration was reduced after HE4 exposure in both cell lysate and culture supernatant (4·8 ± 0·2 to 2·1 ± 0·1 pg/ml in lysate and 7·2 ± 0·3 to 3·6 ± 0·2 pg/ml in supernatant), while the addition of rOPN resulted in a near complete reversal of HE4‐mediated IL‐12 suppression. Similarly, IFN‐Ɣ concentration in cell lysate and supernatant decreased significantly with HE4 (from 35·6 ± 1·0 to 14·4 ± 0·1 pg/ml and from 19·9 ± 0·8 to 11·1 ± 0·6 pg/ml, respectively), and the addition of rOPN again caused recovery of the cytokine levels. As further validation that OPN is the major mediator of HE4‐induced Th1 suppression, treatment of PBMCs with an OPN neutralizing antibody mimicked closely the effect of HE4 on the production of both cytokines (Supporting information, Fig. S4a,b).
Figure 2.
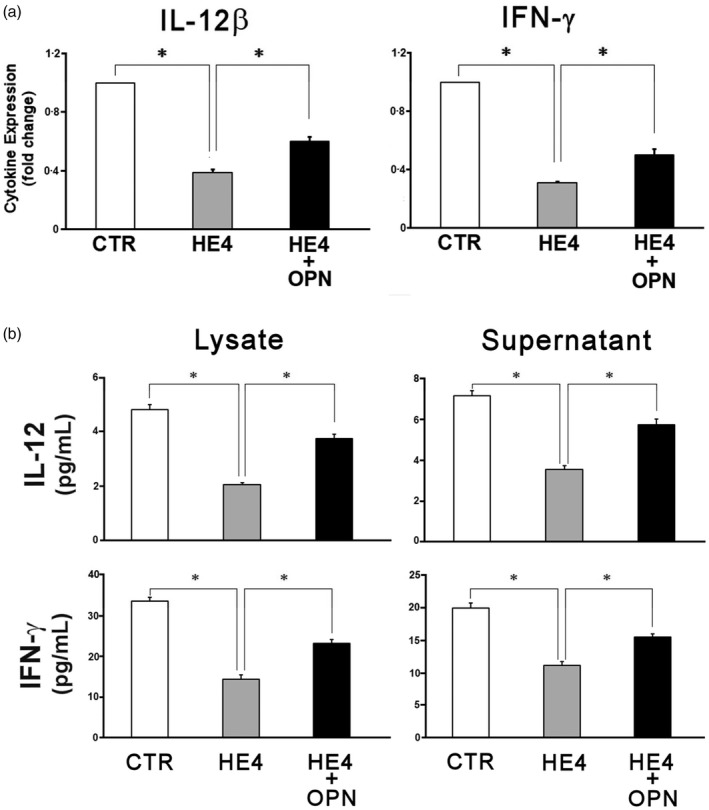
Human epididymis protein 4 (HE4) suppresses expression and secretion of interleukin (IL)‐12 and interferon (IFN)‐Ɣ by peripheral blood mononuclear cells (PBMCs). (a) PBMCs were incubated for 6 h in serum‐free media under the indicated conditions (vehicle, 0·01 μg/ml recombinant HE4 (rHE4) and rHE4 + 20 pg/ml of recombinant osteopontin (rOPN). After 6‐h incubation, transcription levels of IL‐12β (p40) and IFN‐Ɣ were evaluated by real time–polymerase chain reaction (RT–PCR). A bar graph represents relative expression levels against control. (b) The concentrations of IL‐12αβ (p70) and IFN‐Ɣ in the cell lysates and the culture supernatants from 24‐h incubation under the same conditions were measured by enzyme‐linked immunosorbent assay (ELISA). All the quantitative PCRs (qPCRs) and ELISAs were performed with PBMCs from four individual donors. Each assay was repeated four times (qPCR) or 10 times (ELISA). The mean is shown; error bars represent standard error of the mean (s.e.m.). *P < 0·01.
Conditioned media from HE4‐treated PBMCs enhance the viability, proliferation and invasiveness of ovarian cancer cells
In order to assess the effects of PBMC‐produced soluble factors on cancer cell activity, two human ovarian cancer cell lines (SKOV3 and OVCAR8) were incubated with the conditioned media from PBMC cultures (2 × 106 per ml) with rHE4 or vector control. After removal of PBMCs, HE4 was depleted from the media of both culture conditions via immunoprecipitation (Supporting information, Fig. S3). Tumour cells were cultured with HE4‐treated PBMC media showed significantly higher viability at 48 and 72 h (Fig. 3a). Next, a cell migration assay was employed to determine whether conditioned media from rHE4‐exposed PBMCs affects ovarian cancer migration as a surrogate of metastatic capability. The tumour cells that were incubated with HE4‐exposed PBMC media showed more extensive migration than that of control (relative fluorescence units of 1147·2 ± 365·1 versus 3138·1 ± 419·6 for SKOV3 and 1110·1 ± 46·9 versus 4242·2 ± 126·6 for OVCAR8, Fig. 3b). Histocytochemistry using and anti‐Ki67 antibody was performed to evaluate the proliferation of the tumour cells in the presence of rHE4‐exposed PBMC media or vehicle‐exposed conditioned media. The proliferation rate of tumour cells in HE4‐exposed PBMC conditioned media was higher than control media (63·8 ± 18·1 versus 39·9 ± 7·6% for SKOV3 and 62·2 ± 4·5 versus 33·1 ± 3·1 for OVCAR8, Fig. 3c). These findings suggest that PBMCs alter their soluble factor release under the influence of rHE4, thus enhancing the viability, proliferation and migration capabilities of the cultured ovarian cancer cells.
Figure 3.
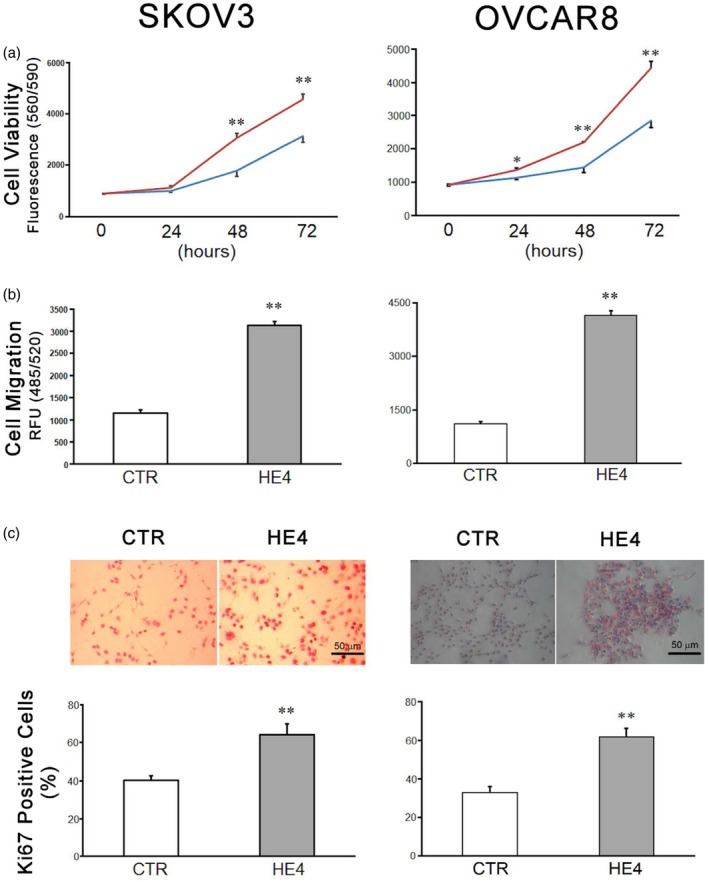
Responses of SKOV3 (left panel) and OVCAR8 (right panel) human ovarian cancer cell lines to the peripheral blood mononuclear cell (PBMC)‐conditioned media. (a) Cells were incubated with conditioned media from the PBMC culture treated with vehicle (blue line) or recombinant human epididymis protein 4 (rHE4) (red line). The cell viabilities were assessed at 24, 48 and 72 h after treatment (n = 10 for 0 h, n = 8 for 24, 48 and 72 h). (b) Cell migration activities with conditioned media were assessed at 24 h of incubation (n = 10). (c) Ki67 immunohistochemistry staining was performed on SKOV3 and OVCAR8 cell lines incubated with the PBMC conditioned media for 24 h. Ki67+ cells are identified with red nuclear staining (upper panel). Bar graph (lower panel) represents the percentage of Ki67+ cells of total countable cells per field under ×200 magnification (n = 6). Scale bar: 50 μm. The mean is shown; error bars represent standard error of the mean (s.e.m.) (n = 10). *P < 0·05; **P < 0·01.
HE4 protects cancer cells from PBMC‐mediated cytotoxicity
In order to evaluate the impact of native (tumour‐cell produced) HE4 on PBMCs, SKOV3 cells were stably transfected with HE4 specific shRNA (shHE4) or scrambled oligonucleotide control plasmid (SO) and then co‐cultured with PBMCs. Clones of shRNA transfected cells were tested for their phenotype by Western blotting and ELISA (Supporting information, Fig. S5). After a 2‐h incubation at 37°C, the effector cells were washed away and the target cells were analysed by flow cytometry. The silencing of HE4 in SKOV3/PBMC co‐cultures led to a significant increase in IL‐12 and IFN‐Ɣ concentrations (Table 2). As shown in Fig. 4, HE4 silencing also increased tumour cell susceptibility to PBMC cytotoxicity, an effect that was reversed by the addition of rHE4. Furthermore, this ‘rescue’ by rHE4 was abrogated at least partially by the addition of rIL‐12 or rIFN‐Ɣ to the culture conditions. These findings suggest that native HE4 production by ovarian cancer cells is critical to cell‐mediated cytotoxicity resistance.
Table 2.
Concentrations of interleukin (IL)‐12, interferon (IFN)‐Ɣ and human epididymis protein 4 (HE4) in co‐culture medium
| Target | Effector | IL‐12 (ng/ml) | INF‐Ɣ (ng/ml) | HE4 (pm) |
|---|---|---|---|---|
| SC | – | – | – | 534·15 ± 41·81 |
| SC | + | 14·84 ± 0·48 | 177·20 ± 1·07 | 639·01 ± 50·38 |
| shHE4 | – | – | – | 174·12 ± 18·55* |
| shHE4 | + | 31·95 ± 0·68** | 417·74 ± 3·54** | 237·91 ± 34·24** |
SC = SKOV3 with scrambled oligo; shHE4 = SKOV3 with HE4 shRNA. Mean ± standard error is shown (n = 10). *P < 0·01 versus subcutaneous (s.c.), **P < 0·01 versus s.c. + effector cells.
Ovarian cancer patient prognosis correlates with the number of intra‐ and peri‐tumoral CD3+ T cells and stromal OPN‐producing cells
Twenty biopsies from high‐grade serous ovarian cancer patients were evaluated by dual fluorescent stain with antibodies against CD3 and OPN (Table 3). Some CD3– tumour cells showed high OPN expression in the tumour segments of the biopsy specimen, while in the stromal area of the biopsy the principal OPN+ cells were CD3+ T cells (Fig. 5a). A significant portion of stromal CD3+ cells possessed strong OPN staining in their cytosols (Fig. 5b). In order to investigate the clinical relationship of HE4, OPN and CD3, numbers of T cells (CD3+) and total OPN+ cells were correlated with preoperative serum HE4 level (available for 13 patients) or PFS duration (available for 16 patients). The numbers of CD3+ infiltrating T cells, both in the tumour and stroma, were directly proportional (tumour, r = 0·541; stroma, r = 0·512) to PFS duration (Fig. 5c). Additionally, the number of OPN+ cells in the tumour and stroma were inversely proportional (tumour; r = –0·635, stroma; r = –0·582) to serum levels of HE4. Moreover, the number of OPN+ cells in the stroma (but not in the tumour) were directly proportional to the patients’ PFS duration (r = 0·711, Fig. 5d). These findings suggest that tissue‐infiltrating, OPN‐expressing T cells play a critical role in the suppression of ovarian cancer progression.
Table 3.
Clinical parameters of donors of biopsies
| pre‐OP sHE4* | PFS† | |
|---|---|---|
| Sample ID | (pmol/ml) | (months) |
| S10‐10110 | 174 | 10 |
| S10‐10726 | n.a. | 16 |
| S10‐15910 | n.a. | 25 |
| S10‐17790 | 376 | 12 |
| S10‐18470 | 529 | 22 |
| S10‐4387 | 462 | 31 |
| S10‐5618 | n.a. | 9 |
| S10‐5842 | n.a. | n.a. |
| S10‐6697 | 150 | n.a. |
| S10‐7183 | 1232 | n.a. |
| S11‐1189 | 3289 | 8 |
| S11‐2223 | 550 | 28 |
| S11‐2493 | 591 | 37 |
| S11‐2684 | 3255 | 24 |
| S11‐3415 | n.a. | n.a. |
| S11‐622 | n.a. | 38 |
| S11‐6675 | 4702 | 16 |
| S11‐6721 | n.a. | 64 |
| S11‐7794 | 410 | 38 |
| S11‐8032 | 623 | 21 |
na, not available.
Pre‐operation serum human epididymis protein 4 (HE4). PFS = progression‐free survival (PFS); n.a. = not available.
Progression‐free survival.
Figure 5.
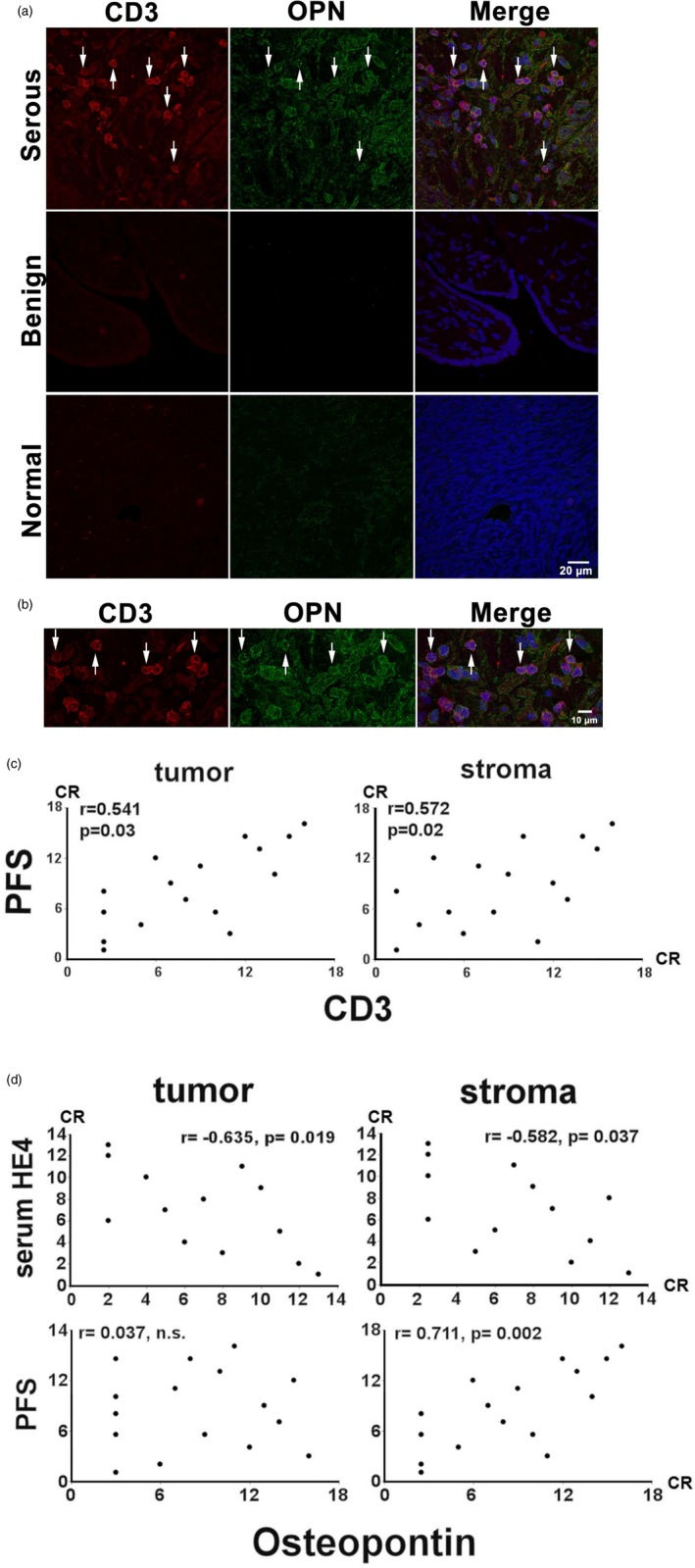
Confocal immunoflurescent analysis of CD3 (DyLight 595, red) and osteopontin (OPN) (DyLight 488, green) expression in biopsy samples. Twenty biopsies (listed in Table 3) were evaluated. (a) Stromal and tumoral CD3+ cells and OPN+ tumour cells are indicated by arrowhead. A biopsy from a benign serous tumour (benign) and an uninvolved section of oophorectomy (normal) were utilized as a negative control. (b). Enlarged image depicting image co‐staining of stromal and tumoral CD3+/OPN+ T cells in their cytosol or the surrounding area (c,d). Graphic representations of Spearman’s rank correlations between the numbers of CD3+ or OPN+ cells and clinical parameters. CR = corrected ranks.
Discussion
HE4 is known to be over‐expressed highly in ovarian cancer, but its causal relationship to ovarian cancer pathogenesis has not been established firmly. Emerging studies suggest that HE4 over‐expression promotes ovarian tumour growth and imparts strong resistance against the most commonly used therapeutics 20, 21, 22, 23, 24. Interestingly, ovarian cancer patients who experienced greater HE4 reduction during neoadjuvant chemotherapy exhibit improved overall survival 24. Our study has shown a novel role for HE4 as a suppressive immunomodulator that acts through OPN inhibition. We have identified the gene for OPN, SPP1, as the most prominently suppressed gene in PBMCs upon HE4 exposure in vitro. Additionally, HE4 was found to down‐regulate OPN production at the transcriptional and protein levels utilizing qPCR and ELISA. As PBMCs are a heterogeneous group of leucocytes, flow cytometric analysis was performed to determine which specific cell type was most impacted by HE4 exposure. We performed staining to differentiate between the four most common players in the PBMC layer: T cells, B cells, NK cells and monocytes. It was determined that T cells (CD3+) experienced a significant reduction in intracellular OPN staining after HE4 expression, while the other cell types showed minimal to no reduction. Although this reduction was relatively modest in comparison to our qPCR and ELISA studies we know that OPN is a heavily secreted cytokine, therefore intracellular staining is likely to under‐represent the true difference in protein expression. To emphasize further the importance of HE4‐mediated OPN suppression in lymphocytes, we found that HE4 exposure resulted in a pronounced reduction in Th1 cytokines IL‐12 and IFN‐Ɣ production by PBMCs, an effect that was reversed partially by the addition of OPN to the culture conditions. It is important to note that, for technical reasons, our subtractive hybridization protocol was performed on the PBMCs of a single donor, and all our in‐vivo studies were performed with leucocytes that were isolated from the blood of healthy donors. It is unclear if normal donor leucocytes are a good surrogate for those of cancer patients that have probably been heavily pre‐exposed to HE4, and a host of other suppressive cytokines. Therefore, we will need to validate these studies further with blood and tissue isolated from ovarian cancer patients.
OPN has historically been considered a pro‐tumorigenic protein. In various types of cancers, serum OPN levels show a direct correlation to tumour burden and an inverse association with patient survival 25, 26, 27. OPN has been suggested to play a critical role in tumour formation and growth by promoting cancer cell survival, proliferation, metastasis and angiogenesis 28, 29. However, other studies have described contrasting anti‐tumour effects of OPN 30, 31, 32, 33. Among these, Crawford et al. used an elegantly designed cancer cell inoculation experiment in OPN null mice to demonstrate that host‐derived OPN can act as a chemoattractant to enhance the host defence activity of macrophages, whereas tumour‐derived OPN inhibits macrophage function, thus promoting tumour escape 30. Building on the theory that OPN possesses dichotomic properties depending on its physical location in relation to the tumour, we have observed that the number of OPN+ cells in the stroma (mainly CD3+ T cells), but not in the tumour (mainly CD3– tumour cells), correlate inversely with patient PFS durations.
To our knowledge, this is the first study to suggest a role for HE4 in ovarian cancer immune escape. We recognize that OPN is known to play a role in humoral immunity 34, 35, 36, which we did not investigate fully here. As we have shown in Table 1 and the Supporting information, Table S1, a variety of genes are modulated in response to HE4 exposure. It is therefore very likely that there are factors other than osteopontin which are also contributing to the inhibitory effect of HE4 on the immune system. Further analysis of the functions of these genes and how they are associated with HE4 is warranted. While we acknowledge the limitations of our study, this work provides the rationale for continued research into the roles of HE4 and osteopontin in ovarian cancer. Upon future validation, we anticipate that the inhibition of HE4 will emerge as a novel treatment strategy for restoring antitumoral immunity in ovarian cancer.
Disclosure
The authors declare no commercial or financial conflicts of interest.
Supporting information
Fig. S1. Human epididymis protein 4 (HE4) reduces peripheral blood mononuclear cell (PBMC) expression of cytokines IL‐12 and IFN‐ɣ, a process that is reversed by osteopontin (OPN) supplementation. PBMCs from four individual donors were incubated for 6 h in serum‐free media with increasing concentrations of recombinant HE4 (rHE4) ranging from 0 to 0·05 μg/ml in the absence (a) or presence (b) of recombinant OPN (rOPN) ranging from 0 to 100 pg/ml. After the incubation, transcriptional levels of interleukin (IL)‐12β (p40) and interferon (IFN)‐Ɣ were evaluated by real‐time polymerase chain reaction (PCR). A bar graph represents relative expression levels against control. The mean is shown; error bars represent standard error of the mean (s.e.m.). *P < 0·05; **P < 0·05 versus 0·01 mg/ml HE4 or 20 pg/ml OPN.
Fig. S2. (a) A diagram of subtractive hybridization. (b) A flowchart of cloning to construct a library.
Fig. S3. Human epididymis protein 4 (HE4) does not impact osteopontin expression in B or natural killer (NK) cells. Two‐colour flow cytometric analysis of peripheral blood mononuclear cells (PBMCs) after a 24‐h incubation with 0·01 μg/ml rHE4 (HE4) or vehicle (CTR). Two‐dimensional (2D)‐scatterplots of osteopontin (OPN) (Alexa Fluor 647) and CD14, CD19 or CD56 [fluorescein isothiocyanate (FITC)] are shown. The mean is shown in the bar graph; error bars represent standard error of the mean (s.e.m.) (n = 3). No substantial differences in OPN expression were observed in CD14+ cells (monocytes), CD19+ cells (B cells) or CD56+ cells (NK cells), although statistical significance was seen in CD14+ cells. *P < 0·05.
Fig. S4. Osteopontin (OPN) neutralization mimics the effect of human epididymis protein 4 (HE4) on peripheral blood mononuclear cell (PBMC) interleukin (IL)‐12 and interferon (IFN)‐Ɣ secretion. PBMCs were incubated in serum‐free media under the indicated conditions (vehicle, 0·01 μg/ml recombinant HE4 (rHE4) and rHE4 + 10 μg/ml of anti‐OPN neutralizing antibody (Ref. no. 1). (a) After a 6‐h incubation, transcriptional levels of IL‐12β (p40) and IFN‐Ɣ were evaluated by real‐time polymerase chain reaction (PCR). A bar graph represents relative expression levels against control. (b) The concentrations of IL‐12αβ(p70) and IFN‐Ɣ in the cell lysates and the culture supernatants from 24‐h incubation were measured by enzyme‐linked immunosorbent assay (ELISA); 10 μg/ml of normal goat immunoglobulin (Ig)G control (R&D Systems) was included in the control (CTR) and HE4 incubations. All the quantitative PCRs (qPCRs) and ELISAs were performed with PBMCs from four individual donors. Each assay was repeated four times (qPCR) or 10 times (ELISA). The mean is shown; error bars represent standard error of the mean (s.e.m.). **P < 0·01.
Fig. S5. Human epididymis protein 4 (HE4) shRNA clones show reduced HE4 production. Western blotting of lysates from SKOV3 cells transfected with shRNA against HE4. Cell lysates were obtained from quiescent cells; 50 μg/lane of proteins were run on sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS‐PAGE) and immunoblotted with rabbit anti‐HE4 polyclonal antibody (Abcam; ab109298). An image of gel stained after transfer was shown as a loading monitor. (b) The bar graph represents the concentrations of HE4 in culture supernatant (SN) and cell lysates (lysate) from each clone; 1 × 106 cells/well of a six‐well plate were incubated in serum‐free media for 24 h. Appropriately diluted culture supernatants or 2·5 μg/μl of lysate proteins were served for HE4 ELISA. shHE4 clone 5 was used as an HE4 silenced SKOV3 cell in the study. SO = scrambled oligo‐transfected control clone. The mean is shown; error bars represent standard error of the mean (s.e.m.) (n = 10).
Table S1. Frequency and categories of differentially expressed genes in peripheral blood mononuclear cells (PBMCs) in response to human epididymis protein 4 (HE4)
Table S2. Summary of polymerase chain reaction (PCR) primer sequences
Table S3. Human epididymis protein 4 (HE4) concentrations in conditioned media
Acknowledgement
This work is supported partially by a grant from the National Heart, Lung, and Blood Institute of National Institute of Health Grant (R01 HL089405 and R01 HL115265).
References
- 1. Kirchhoff C, Habben I, Ivell R, Krull N. A major human epididymis‐specific cDNA encodes a protein with sequence homology to extracellular proteinase inhibitors. Biol Reprod 1991;45:350–7. [DOI] [PubMed] [Google Scholar]
- 2. Ranganathan S, Simpson KJ, Shaw DC, Nicholas KR. The whey acidic protein family: a new signature motif and three‐dimensional structure by comparative modeling. J Mol Graph Model 1999;17:106–13. [DOI] [PubMed] [Google Scholar]
- 3. Schalkwijk J, Wiedow O, Hirose S. The trappin gene family: proteins defined by an N‐terminal transglutaminase substrate domain and a C‐terminal four‐disulphide core. Biochem J 1999;340:569–77. [PMC free article] [PubMed] [Google Scholar]
- 4. Bingle L, Singleton V, Bingle CD. The putative ovarian tumour marker gene HE4 (WFDC2), is expressed in normal tissues and undergoes complex alternative splicing to yield multiple protein isoforms. Oncogene 2002;21:2768–73. [DOI] [PubMed] [Google Scholar]
- 5. Galgano MT, Hampton GM, Frierson HF Jr. Comprehensive analysis of HE4 expression in normal and malignant human tissues. Mod Pathol 2006;19:847–53. [DOI] [PubMed] [Google Scholar]
- 6. Drapkin R, von Horsten HH, Lin Y et al Human epididymis protein 4 (HE4) is a secreted glycoprotein that is overexpressed by serous and endometrioid ovarian carcinomas. Cancer Res 2005;65:2162–9. [DOI] [PubMed] [Google Scholar]
- 7. Schaner ME, Ross DT, Ciaravino G et al Gene expression patterns in ovarian carcinomas. Mol. Biol Cell 2003;14:4376–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hellström I, Raycraft J, Hayden‐Ledbetter M et al The HE4 (WFDC2) protein is a biomarker for ovarian carcinoma. Cancer Res 2003;63:3695–700. [PubMed] [Google Scholar]
- 9. Angioli R, Capriglione S, Aloisi A et al Can HE4 predict platinum response during first‐line chemotherapy in ovarian cancer? Tumour Biol 2014;35:7009–15. [DOI] [PubMed] [Google Scholar]
- 10. Chudecka‐Głaz AM, Cymbaluk‐Płoska AA, Menkiszak JL, Sompolska‐Rzechuła AM, Tołoczko‐Grabarek AI, Rzepka‐Górska IA. Serum HE4, CA125, YKL‐40, bcl‐2, cathepsin‐L and prediction optimal debulking surgery, response to chemotherapy in ovarian cancer. J. Ovarian. Res 2014;7:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vallius T, Hynninen J, Auranen A et al Serum HE4 and CA125 as predictors of response and outcome during neoadjuvant chemotherapy of advanced high‐grade serous ovarian cancer. Tumour Biol 2014;35:12389–95. [DOI] [PubMed] [Google Scholar]
- 12. Moore RG, Brown AK, Miller MC et al The use of multiple novel tumor biomarkers for the detection of ovarian carcinoma in patients with a pelvic mass. Gynecol Oncol 2008;108:402–8. [DOI] [PubMed] [Google Scholar]
- 13. Moore RG, Brown AK, Miller MC et al Utility of a novel serum tumor biomarker HE4 in patients with endometrioid adenocarcinoma of the uterus. Gynecol Oncol 2008;110:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moore RG, McMeekin DS, Brown AK et al A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol Oncol 2009;112:40–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moore RG, Jabre‐Raughley M, Brown AK et al Comparison of a novel multiple marker assay vs the Risk of Malignancy Index for the prediction of epithelial ovarian cancer in patients with a pelvic mass. Am J Obstet Gynecol 2010;203(228):e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Denhardt DT, Lopez CA, Rollo EE, Hwang SM, An XR, Walther SE. Osteopontin‐induced modifications of cellular functions. Ann NY Acad Sci 1995;760:127–42. [DOI] [PubMed] [Google Scholar]
- 17. O’Regan AW, Chupp GL, Lowry JA, Goetschkes M, Mulligan N, Berman JS. Osteopontin is associated with T cells in sarcoid granulomas and has T cell adhesive and cytokine‐like properties in vitro. J Immunol 1999;162:1024–31. [PubMed] [Google Scholar]
- 18. Ashkar S, Weber GF, Panoutsakopoulou V et al Eta‐1 (Osteopontin): an early component of type‐1 (cell‐mediated) immunity. Science 2000;287:860–4. [DOI] [PubMed] [Google Scholar]
- 19. Huhtinen K, Suvitie P, Hiissa J et al Serum HE4 concentration differentiates malignant ovarian tumours from ovarian endometriotic cysts. Br J Cancer 2009;100:1315–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ribeiro JR, Schorl C, Yano N et al HE4 promotes collateral resistance to cisplatin and paclitaxel in ovarian cancer cells. J Ovarian Res 2016;9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moore RG, Hill EK, Horan T et al HE4 (WFDC2) gene overexpression promotes ovarian tumor growth. Sci Rep 2014;4:3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pelissier A, Roulot A, Guéry B, Bonneau C, Bellet D, Rouzier R. Serum CA125 and HE4 levels as predictors for optimal interval surgery and platinum sensitivity after neoadjuvant platinum‐based chemotherapy in patients with advanced epithelial ovarian cancer. J Ovarian Res 2016;9:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee S, Choi S, Lee Y, Chung D, Hong S, Park N. Role of human epididymis protein 4 in chemoresistance and prognosis of epithelial ovarian cancer. J Obstet Gynecol Res 2017;43:220–7. [DOI] [PubMed] [Google Scholar]
- 24. Plotti F, Scaletta G, Capriglione S et al The role of HE4, a novel biomarker, in predicting optimal cytoreduction after neoadjuvant chemotherapy in advanced ovarian cancer. Int J Gynecol Cancer 2017;27:696–702. [DOI] [PubMed] [Google Scholar]
- 25. Singhal H, Bautista DS, Tonkin KS et al Elevated plasma osteopontin in metastatic breast cancer associated with increased tumor burden and decreased survival. Clin Cancer Res 1997;3:605–11. [PubMed] [Google Scholar]
- 26. Chiodoni C, Colombo MP, Sangaletti S. Matricellular proteins: from homeostasis to inflammation, cancer, and metastasis. Cancer Metastasis Rev 2010;29:295–307. [DOI] [PubMed] [Google Scholar]
- 27. Kluger HM, Hoyt K, Bacchiocchi A et al Plasma markers for identifying patients with metastatic melanoma. Clin Cancer Res 2011;17:2417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bellahcène A, Castronovo V, Ogbureke KU, Fisher LW, Fedarko NS. Small integrin‐binding ligand N‐linked glycoproteins (SIBLINGs): multifunctional proteins in cancer. Nat Rev Cancer 2008;8:212–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chong HC, Tan CK, Huang RL, Tan NS. Matricellular proteins: a sticky affair with cancers. J Oncol 2012;2012:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Crawford HC, Matrisian LM, Liaw L. Distinct roles of osteopontin in host defense activity and tumor survival during squamous cell carcinoma progression in vivo . Cancer Res 1998;58:5206–15. [PubMed] [Google Scholar]
- 31. Bourassa B, Monaghan S, Rittling SR. Impaired anti‐tumor cytotoxicity of macrophages from osteopontin‐deficient mice. Cell Immunol 2004;227:1–11. [DOI] [PubMed] [Google Scholar]
- 32. Hsieh YH, Margaret Juliana M, Ho KJ et al Host‐derived osteopontin maintains an acute inflammatory response to suppress early progression of extrinsic cancer cells. Int J Cancer 2012;131:322–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Danzaki K, Kanayama M, Alcazar O, Shinohara ML. Osteopontin has a protective role in prostate tumor development in mice. Eur J Immunol 2016;46:2669–78. [DOI] [PubMed] [Google Scholar]
- 34. Take Y, Nakata K, Hashimoto J et al Specifically modified osteopontin in rheumatoid arthritis fibroblast‐like synoviocytes supports interaction with B cells and enhances production of interleukin‐6. Arthritis Rheum 2009;60:3591–601. [DOI] [PubMed] [Google Scholar]
- 35. Guo B, Tumang JR, Rothstein TL. B cell receptor crosstalk: B cells express osteopontin through the combined action of the alternate and classical BCR signaling pathways. Moll Immunol 2009;46:587–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kon S, Nakayama Y, Matsumoto N et al A novel cryptic binding motif, LRSKSRSFQVSDEQY, in the C‐terminal fragment of MMP‐3/7‐cleaved osteopontin as a novel ligand for α9β1 integrin is involved in the anti‐type II collagen antibody‐induced arthritis. PLOS ONE 2014;9:e116210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Human epididymis protein 4 (HE4) reduces peripheral blood mononuclear cell (PBMC) expression of cytokines IL‐12 and IFN‐ɣ, a process that is reversed by osteopontin (OPN) supplementation. PBMCs from four individual donors were incubated for 6 h in serum‐free media with increasing concentrations of recombinant HE4 (rHE4) ranging from 0 to 0·05 μg/ml in the absence (a) or presence (b) of recombinant OPN (rOPN) ranging from 0 to 100 pg/ml. After the incubation, transcriptional levels of interleukin (IL)‐12β (p40) and interferon (IFN)‐Ɣ were evaluated by real‐time polymerase chain reaction (PCR). A bar graph represents relative expression levels against control. The mean is shown; error bars represent standard error of the mean (s.e.m.). *P < 0·05; **P < 0·05 versus 0·01 mg/ml HE4 or 20 pg/ml OPN.
Fig. S2. (a) A diagram of subtractive hybridization. (b) A flowchart of cloning to construct a library.
Fig. S3. Human epididymis protein 4 (HE4) does not impact osteopontin expression in B or natural killer (NK) cells. Two‐colour flow cytometric analysis of peripheral blood mononuclear cells (PBMCs) after a 24‐h incubation with 0·01 μg/ml rHE4 (HE4) or vehicle (CTR). Two‐dimensional (2D)‐scatterplots of osteopontin (OPN) (Alexa Fluor 647) and CD14, CD19 or CD56 [fluorescein isothiocyanate (FITC)] are shown. The mean is shown in the bar graph; error bars represent standard error of the mean (s.e.m.) (n = 3). No substantial differences in OPN expression were observed in CD14+ cells (monocytes), CD19+ cells (B cells) or CD56+ cells (NK cells), although statistical significance was seen in CD14+ cells. *P < 0·05.
Fig. S4. Osteopontin (OPN) neutralization mimics the effect of human epididymis protein 4 (HE4) on peripheral blood mononuclear cell (PBMC) interleukin (IL)‐12 and interferon (IFN)‐Ɣ secretion. PBMCs were incubated in serum‐free media under the indicated conditions (vehicle, 0·01 μg/ml recombinant HE4 (rHE4) and rHE4 + 10 μg/ml of anti‐OPN neutralizing antibody (Ref. no. 1). (a) After a 6‐h incubation, transcriptional levels of IL‐12β (p40) and IFN‐Ɣ were evaluated by real‐time polymerase chain reaction (PCR). A bar graph represents relative expression levels against control. (b) The concentrations of IL‐12αβ(p70) and IFN‐Ɣ in the cell lysates and the culture supernatants from 24‐h incubation were measured by enzyme‐linked immunosorbent assay (ELISA); 10 μg/ml of normal goat immunoglobulin (Ig)G control (R&D Systems) was included in the control (CTR) and HE4 incubations. All the quantitative PCRs (qPCRs) and ELISAs were performed with PBMCs from four individual donors. Each assay was repeated four times (qPCR) or 10 times (ELISA). The mean is shown; error bars represent standard error of the mean (s.e.m.). **P < 0·01.
Fig. S5. Human epididymis protein 4 (HE4) shRNA clones show reduced HE4 production. Western blotting of lysates from SKOV3 cells transfected with shRNA against HE4. Cell lysates were obtained from quiescent cells; 50 μg/lane of proteins were run on sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS‐PAGE) and immunoblotted with rabbit anti‐HE4 polyclonal antibody (Abcam; ab109298). An image of gel stained after transfer was shown as a loading monitor. (b) The bar graph represents the concentrations of HE4 in culture supernatant (SN) and cell lysates (lysate) from each clone; 1 × 106 cells/well of a six‐well plate were incubated in serum‐free media for 24 h. Appropriately diluted culture supernatants or 2·5 μg/μl of lysate proteins were served for HE4 ELISA. shHE4 clone 5 was used as an HE4 silenced SKOV3 cell in the study. SO = scrambled oligo‐transfected control clone. The mean is shown; error bars represent standard error of the mean (s.e.m.) (n = 10).
Table S1. Frequency and categories of differentially expressed genes in peripheral blood mononuclear cells (PBMCs) in response to human epididymis protein 4 (HE4)
Table S2. Summary of polymerase chain reaction (PCR) primer sequences
Table S3. Human epididymis protein 4 (HE4) concentrations in conditioned media


