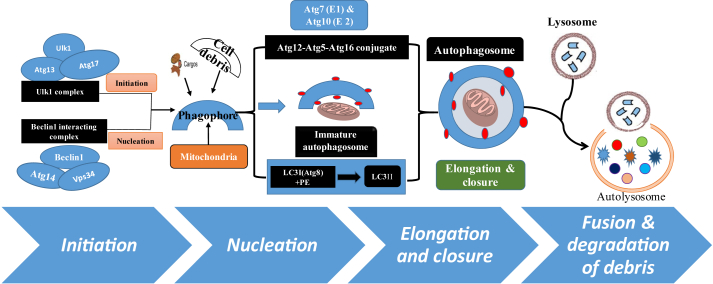
Figure 1.
Schematic diagram showing the overview of basic and molecular processes involved in autophagy. The process of autophagy involves a series of dynamic membrane rearrangements regulated by a set of ATG proteins, which can be modulated by the post-translational modifications (re-review in ref. 52, 53 and 70). In brief, it involves the following key steps i) control of phagophore formation by beclin-1/VPS34 at the ER and other membranes in response to stress such as starvation or diabetes; ii) Atg5–Atg12 conjugation, interaction with Atg16L and polymerization at the immature phagophore; iii) the LC3 processing and insertion into the extending phagophore membrane; iv) then engulfing of random or selective targets for degradation, completion of the autophagosome and recycling of some LC3-II/ATG8 v) and finally fusion of the autophagosome with the lysosome and proteolytic degradation by lysosomal proteases of engulfed molecules or cellular debris.