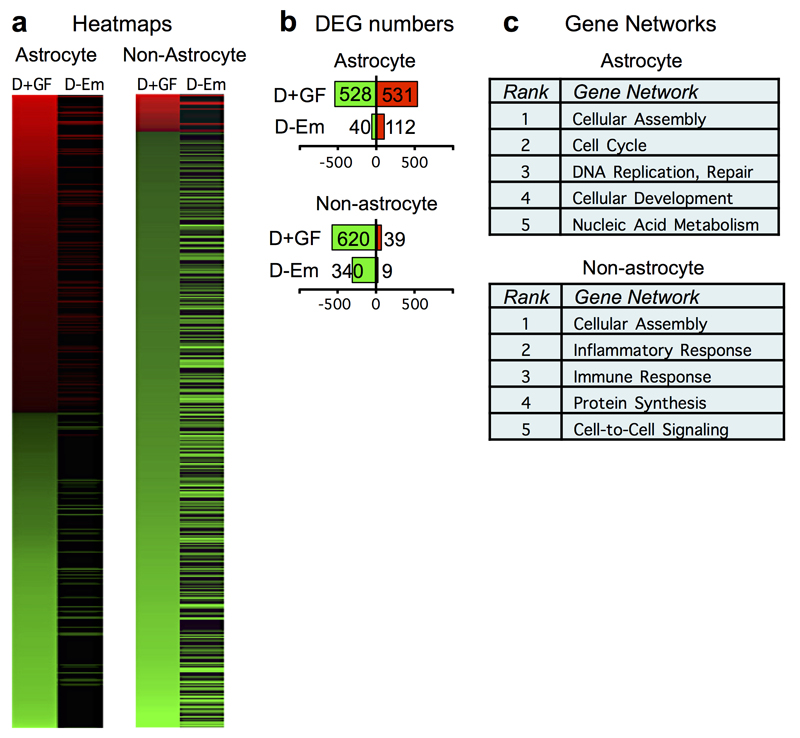
Extended Data Figure 7. Comparison of genomic data from astrocytes and non-astrocyte cells from mice with or without FGF+EGF after SCI.
a, Heat maps showing significantly differentially expressed genes (DEG) derived by RNA-Seq of mRNAs from spinal cord tissue of mice treated with SCI+1D+FGF+EGF (D+GF), and the expression of these genes in mice treated with SCI+1D-empty (D-Em), at two weeks after SCI. Data are shown for mRNAs derived selectively from astrocytes or from all other cell types (non-astrocytes), isolated as previously described19. Red upregulated, green downregulated relative to SCI-only. (n = 3 mice per group. FDR<0.1 for differential expression). b, Total numbers of significant DEGs in astrocytes and non-astrocytes from mice as shown in heatmap in panel a. Red and green numerical values indicate significantly upregulated and downregulated genes, respectively. Relative to SCI-only, over 900 astrocyte genes and over 300 non-astrocyte genes were significantly up- or down-regulated in mice with 12 days of growth factor treatment, which were not significantly altered by treatment with empty depots. c, Top five networks of genes significantly altered by D+GF that were not altered by D–empty after SCI relative to SCI-only, as identified by unbiased analysis (Ingenuity ®). RNAseq data are available at NCBI’s Gene Expression Omnibus repository (GSE111529).