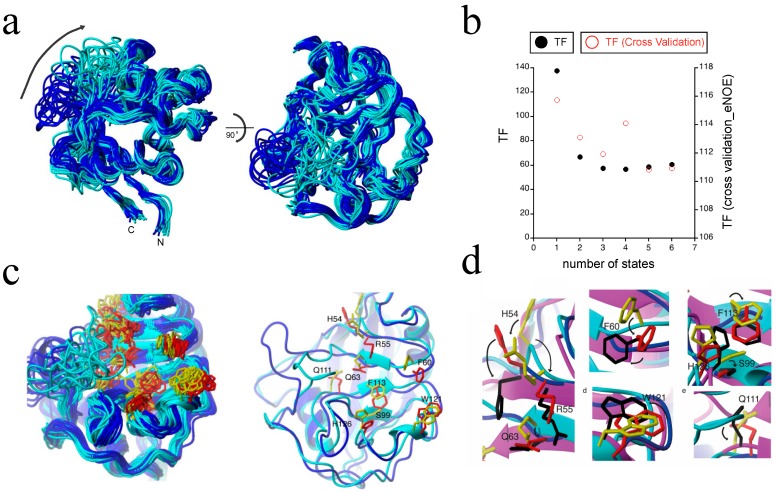
Figure 8.
The eNOE detection of an allosteric network in cyclophilin A. (a) A structural ensemble of cyclophilin A in its apo form is shown, highlighting the presence of two distinct states. Each of the 20 conformers represents two states. States were color-coded as open (blue) and closed (cyan). Two distinct states are observed throughout most of the structure. The orientation of the structural ensemble shown on the right is a 90° rotation of that on the left; (b) The dependence of the CYANA TF (black) and the overall TF from the jackknife-type cross-validation (red) are shown as functions of the number of states; (c) The two-state ensemble of the active site residues of cyclophilin A. On the left, a ribbon representation of the 20 conformers is shown, color-coded individually for the two states: the closed state is shown in cyan for the backbone and in yellow for the side chains of the active site, while the open state is shown in blue for the backbone and in red for the side chains in the active site. The right shows a single representative from each state, with the side chains labeled. The lowest energy two-state conformers were selected; (d) Proposed mechanism of action of cyclophilin A at the atomic resolution. The X-ray structure of cyclophilin A in complex with the HIV-1 capsid protein (PDB ID: 1ak4 [59]) was superimposed with the presented two-state ensemble, which highlights the fact that the open-state matched the ligand-bound state well. The closed state is shown in cyan for the backbone ribbon and in yellow for the side chains, the open state in blue for the backbone ribbon and in red for the side chains, and the X-ray structure is shown in purple for the backbone ribbon and in black for the side chains. Individual close-ups of the superposition are shown. The potential modes of action for catalysis of the individual residues are indicated by arrows. Reprinted from [36], Copyright (2015), with permission from Wiley.