Figure 1.
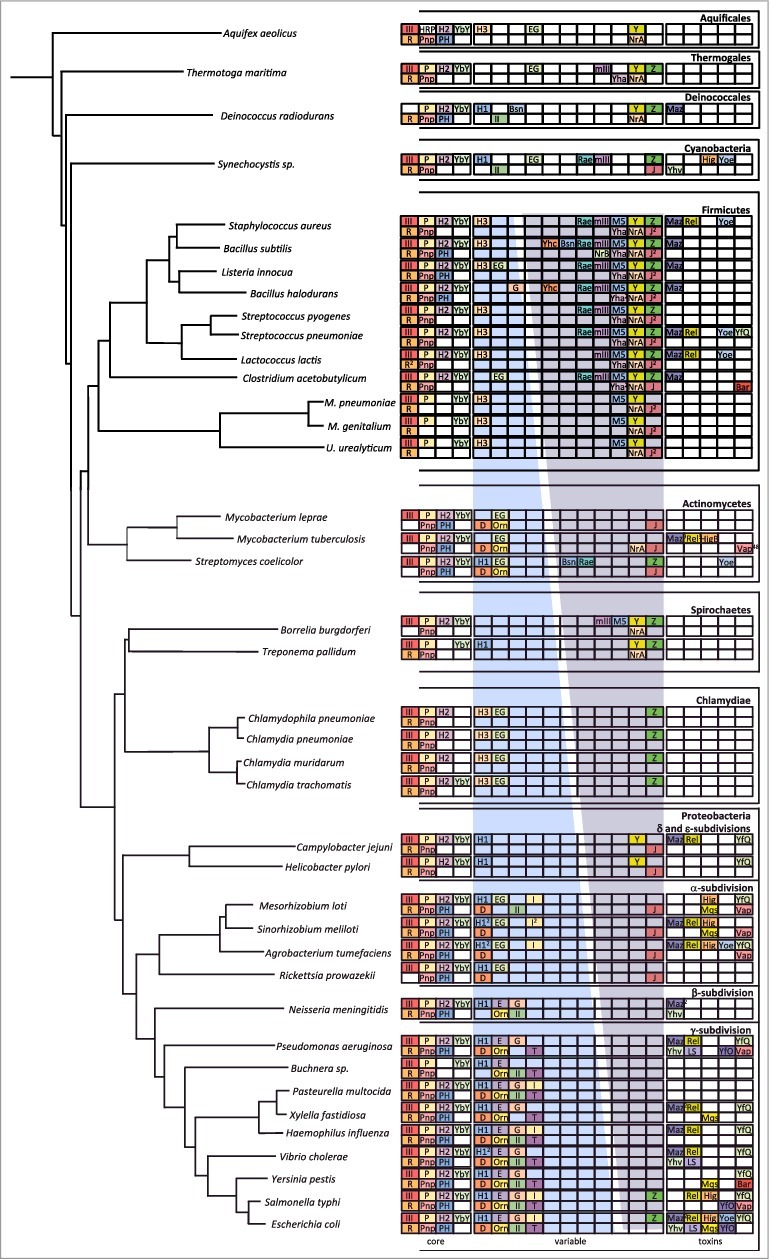
Phylogeny of ribonucleases in bacteria. The image is an update of Figure 1 in [ref. 18]. The most highly conserved RNases are in a subsection labelled ‘core’. More variable RNases are shown in a subsection labelled ‘variable’. For these two subsections, endoribonucleases are confined to the top line and exoribonucleases to the bottom line. Within the variable subsection, the blue gradient represents the distribution of RNases mostly identified with the E. coli degradation machinery; the purple gradient represents the distribution of RNases mostly identified in B. subtilis. Horizontally transferred toxin RNases (all endoribonucleases) are shown in a subsection labelled ‘toxins’. Abbreviations are as follows: Endondonucleases: III = RNase III; P = RNase P; H2 = RNase HII; H1 = RNase HI; H3 = RNase HIII; E = RNase E; G = RNase G; EG = RNase E/G; YbY = YbeY; I = RNase I; Z = RNase Z; M5 = RNase M5; Y = RNase Y; mIII = Mini-III; Rae = Rae1; Maz = MazF; Rel = RelE; Yoe = YoeB; Hig = HigB; YfQ = YafQ; Yhv = YhaV; LS = RNase LS, Mqs = MqsR; YfO = YafO; Bsn = RNase Bsn; Bar = Barnase; Exoribonucleases: R = RNase R; Pnp = polynucleotide phosphorylase; PH = RNase PH; D = RNase D; Orn = oligoribonuclease; II = RNase II; T = RNase T; J = RNase J; BN = RNase BN; Yha = RNase YhaM. Numbers in superscript refer to the number of orthologs present when identified.
