Figure 1.
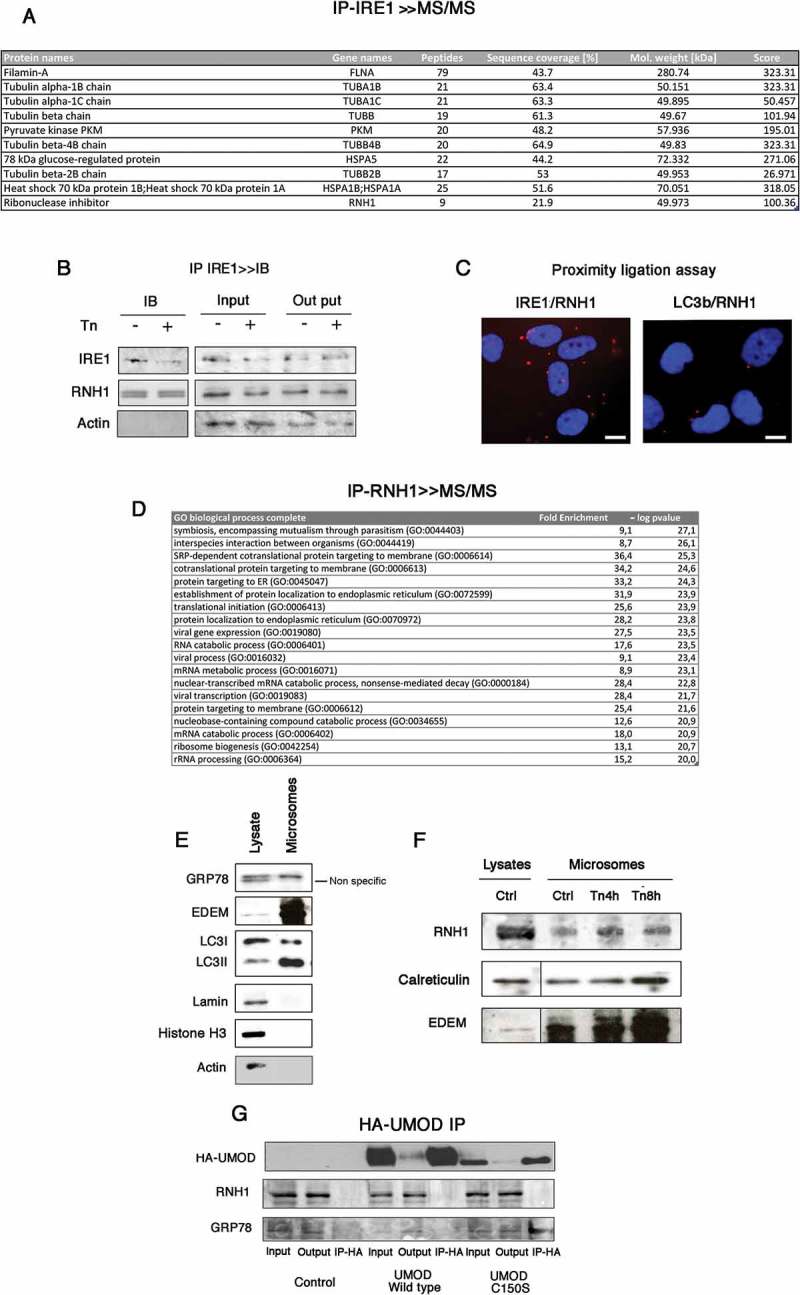
RNH1 interacts with IRE1 at the surface of the ER. A. Endogenous IRE1 was immunoprecipitated (IP) followed by tandem mass spectrometry. The IPs presented were processed as described in the Materials and Methods and analyzed by tandem mass spectrometry. Peptides identified for RNH1 and proteins known to interact with IRE1 in the analysis are indicated. B. Endogenous interaction between RNH1 and IRE1 were analyzed by immunoblotting after the IP of IRE1. Cells were incubated or not with 2.5 μg/l tunicamycin for 4 h. IRE1 was immunoprecipitated and analyzed by western blot to detect RNH1 and IRE1 expression. Data are representative of 3 independent experiments. Input refers to the lysate before IP, and output to the lysate after IP. C. Proximity ligation assay was performed using antibodies directed against IRE1, LC3b and RNH1. Red dots indicate proximity between IRE1 and RNH1 of less than 40 nm. Bar represents 10 μm D. Endogenous RNH1 was immunoprecipitated (IP), followed by tandem mass spectrometry. Proteins were annotated using the Gene Ontology Biological process, and enrichments analyses were performed using the PANTHER Overrepresentation Test on a list of 144 proteins identified in 3 independent experiments. The GO terms in red are associated with a p value < 10−20 after Bonferroni correction for multiple testing. E. Western-blot analysis of microsomes preparations and whole cell lysate for the expression of ER markers, nucleus marker and cytoplasmic markers. F. Western-blot analysis of microsome preparations and whole cell lysate for the expression of ER markers, nucleus marker and cytoplasmic markers. G. Cells were incubated or not with 2.5 μg/l tunicamycin for up to 8 h and then microsomes and protein extracts were prepared. RNH1, Calreticulin and EDEM expression in cell lysates and microsomes was analyzed by western blotting. Data are representative of 3 independent experiments. H. Cells were transfected with an expression vector for UMOD wild type-HA or mutated UMOD C150S or empty vector. Western blots for GRP78 and RNH1 were performed on IP-HA. Data are representative of 3 independent experiments.
