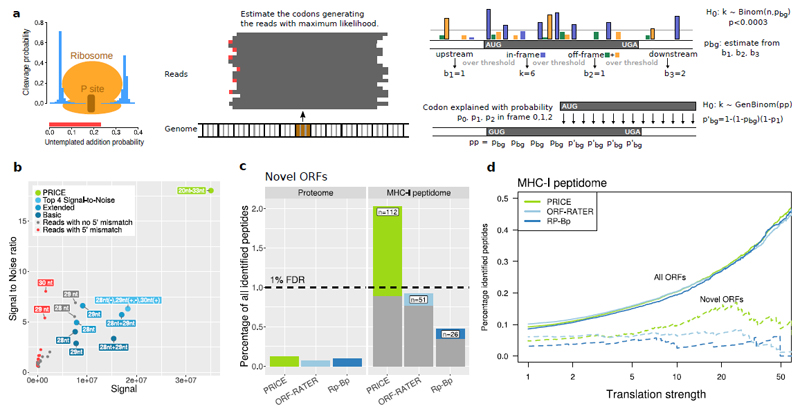
Figure 1. The PRICE approach.
(a) Schematic of the approach. Left: Bars represent parameters of the probabilistic model. Center: Translated codons are identified by solving the inverse problem of the model. Right: calling actively translated ORFs based on the generalized binomial distribution (for details see Online Methods).
(b) Approaches to map reads to codons are compared with respect to signal (total number of reads mapped in-frame) and the signal to noise ratio (noise: reads mapped out-of-frame to annotated ORFs). Colors represent deterministic mapping of read classes defined by length and 5’ mismatch state (red, grey), of combinations of read classes (blue; Basic: ignoring 5’ mismatches; Extended: considering 5’ mismatches; Top 4: combining the best read classes; see also Supplementary Fig. 4) and probabilistic mapping by PRICE (green).
(c) Total amount of peptides detected in proteome and MHC-I peptidome mass spectrometry experiments (MHC-I peptidome data set 1; see Supplementary Fig. 9a for the other experiment). The 1% peptide identification FDR is indicated. Grey bars show the peptides from ORFs also identified by ORF-RATER or Rp-Bp (for PRICE) or ORFs also identified by PRICE (for ORF-RATER and Rp-Bp).
d) Validation rates of peptides from predicted ORFs with a minimal number of reads per codon (MHC-I peptidome data set 2; see Supplementary Fig. 9b for the other experiment). Rates for all ORFs (solid lines) identified by the indicated methods and for ORFs predicted de-novo (dashed lines) are shown.