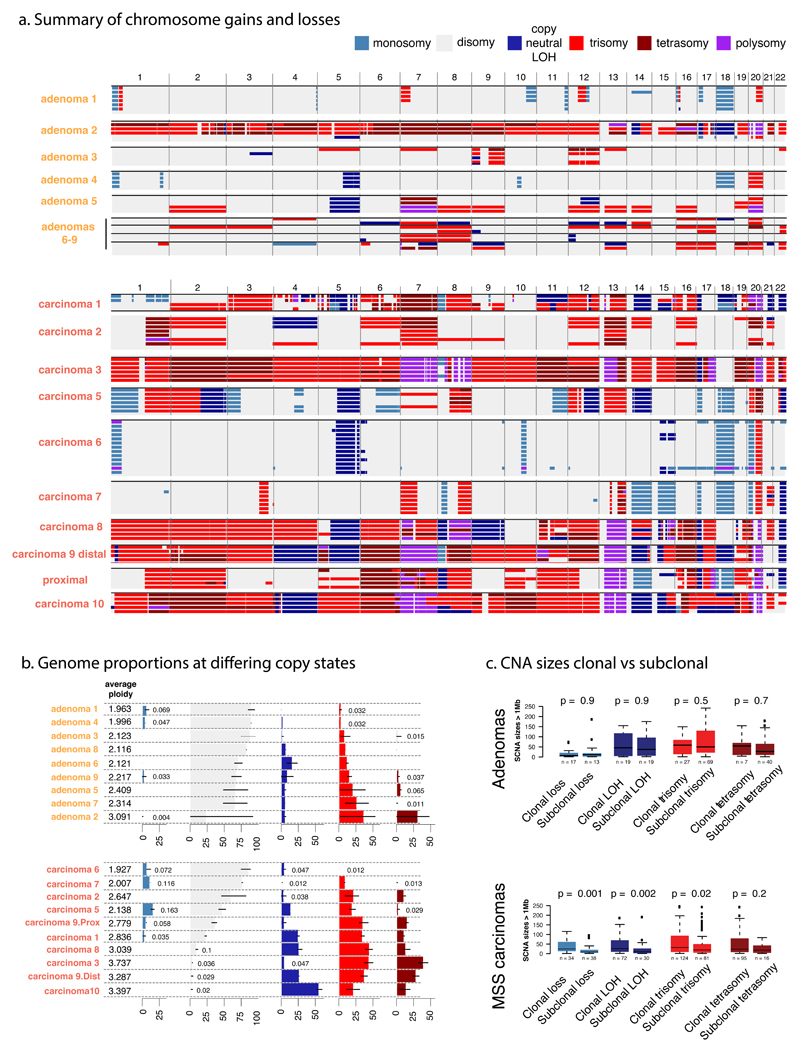
Figure 3. Copy number alterations in CRAs and MSS CRCs.
a. A genome-wide view of CNAs is shown for each region of CRAs (top) and CRCs (bottom). Cancers show a greater CNA burden than adenomas, and most CNAs are clonal in cancers, whereas CRAs show more frequent sub-clonal CNAs. Copy number ≥5 is shown as “polysomy”. b. The figure shows estimated ploidy and summarises the proportion of each tumour at different copy-states. Black bars show the range of biopsy copy-numbers. c. Size distributions of ubiquitous and sub-clonal (branch and leaf) CNAs demonstrate the preference of CRCs to have larger events. Boxplots show the median and inter quantile range (IQR), upper whisker is 3rd quantile + 1.5*IQR and lower whisker is 1st quantile - 1.5*IQR. The colour-coding of copy number states (top right) applies to all panels.