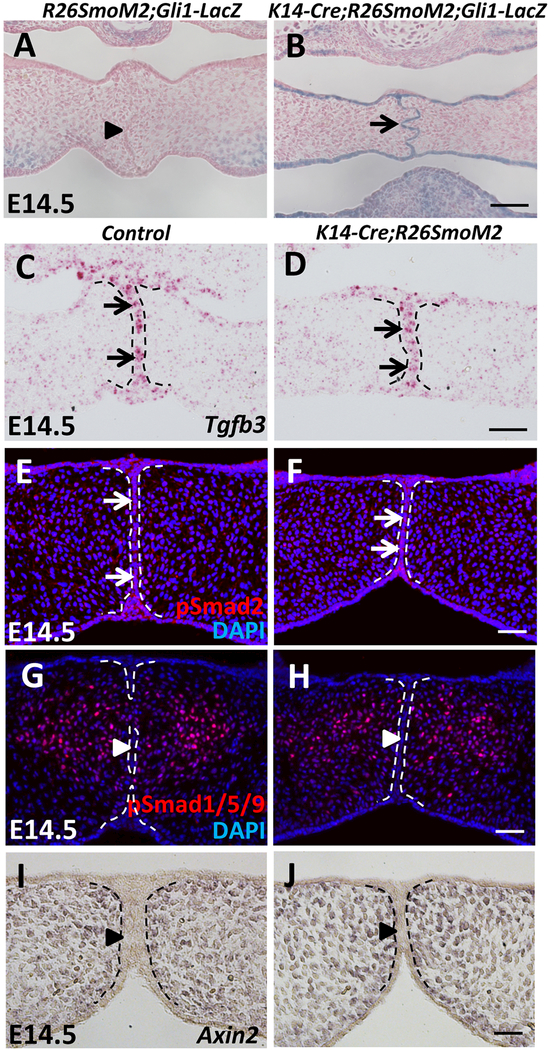
Figure 2. The Hh pathway is constitutively activated in the MEE of SmoM2 mutant mice without affecting the Tgf-β, Smad-dependent Bmp, or canonical Wnt pathways.
(A-B) X-gal staining (blue) of frontal sections of E14.5 R26SmoM2;Gli1-LacZ (control) and K14-Cre;R26SmoM2;Gli1-LacZ palatal shelves. Arrow indicates activated Gli1 expression, whereas arrowhead indicates absence of Gli1 expression. (C-D) RNAscope in situ hybridization analysis of Tgfb3 (red dots, indicated by arrows) in frontal sections of E14.5 control and K14-Cre;R26SmoM2 palatal shelves. (E-H) Immunofluorescence of phospho-Smad2 (pSmad2) and phospho-Smad1/5/9 (pSmad1/5/9) (red) in frontal sections of E14.5 control and K14-Cre;R26SmoM2 palatal shelves. Arrows indicate positive signal; arrowheads indicate absence of signal. (I-J) In situ hybridization of Axin2 (dark blue) in frontal sections of E14.5 control and K14-Cre;R26SmoM2 palatal shelves. Arrowheads indicate absence of Axin2 expression. Black and white dotted lines indicate the location of the MEE. Scale bars: 50 μm.