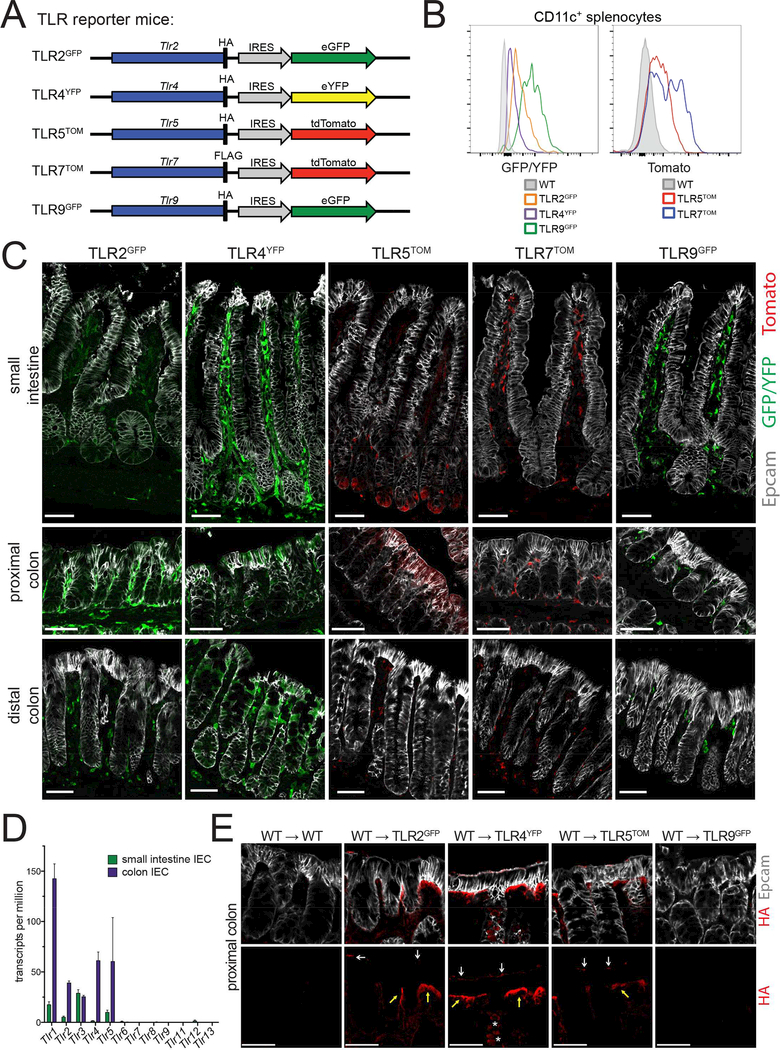
Figure 1: Epithelial TLR expression differs dramatically between the SI and colon.
(A)Design of TLR reporter mice. A reporter cassette including an epitope tag (HA or FLAG) and an internal ribosome entry site followed by a fluorescent protein (eGFP, eYFP, or tdTomato) was inserted at the 3’ end of the endogenous TLR gene for the TLRs indicated. Note: for simplicity, the exon/intron structure for each TLR is not shown.
(B) Expression of TLR reporter genes was confirmed by measuring fluorescence (GFP/YFP or tomato) by flow cytometry from live, CD11c+ cells from the spleens of indicated mice. Flow cytometry plots are representative of 2 independent experiments, each including at least 3 mice of each genotype.
(C) SI and colon sections of reporter mice stained with Epcam (epithelial cells) and antibodies to either GFP/YFP or tomato (TLR reporters). All SI images were from the ileum. Images were taken at 20× magnification and exposures were set using staining of WT mouse tissues. Images are representative of at least 3 independent experiments, each including 2–5 mice of each genotype. Scale bars are 50 microns.
(D) TLR expression in IECs was assessed by RNA-sequencing performed on IECs isolated from WT mice from 2 litters. Values are displayed as a number of transcripts per million total reads. Error bars are mean +/− SEM.
(E) Proximal colon sections from radiation chimeras in which WT bone marrow was reconstituted into WT or TLR reporter mice stained with Epcam (epithelial cells) and antibodies to the HA epitope tag (TLR protein). HA staining is separated in bottom panels for easier visualization. Images were taken at 20× magnification and exposures were set using staining of WT mouse tissues. White arrows indicate apical staining, yellow arrows indicate basolateral staining, and stars indicate intracellular staining. Images are representative of 2 independent experiments, each including 1–4 mice of each genotype. Scale bars are 50 microns.