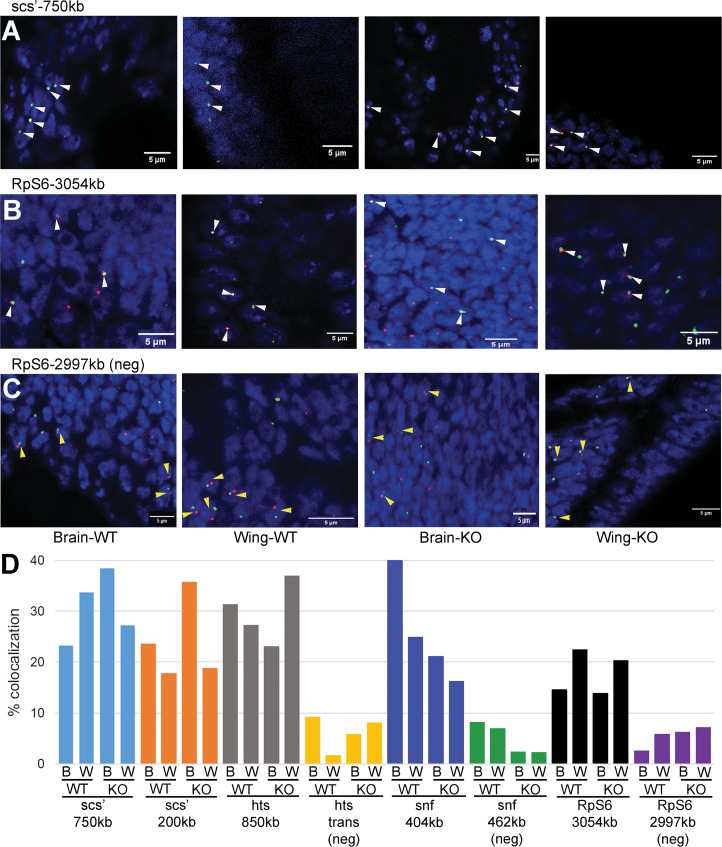
Fig 4. Testing 4C interactions by FISH.
Representative confocal micrographs of hybridizations to third instar larval brains and wing discs from BEAF wild-type (WT) or null (KO; BEAFAB-KO) animals are shown, as labeled. DNA is labeled with DAPI to identify nuclei. White and yellow arrowheads indicate representative nuclei with colocalizing or non-colocalizing FISH signals, respectively. (A) Hybridization to the scs’ viewpoint (green) and a sequence around 750 kb away (red). Overlapping signals are yellow. (B) Hybridization to the RpS6 viewpoint (green) and a sequence around 3054 kb away (red). (C) Negative control hybridization to the RpS6 viewpoint (green) and a sequence around 2997 kb away (red) that does not represent a 4C interaction. (D) Graph of all FISH results, indicating % of counted nuclei showing colocalization of signals (n = 131 to 525, see S6 Table). The viewpoint probe and distance to the second probe are indicated, as well as if the second probe was a negative control (neg) that did not represent a 4C interaction. W: third instar larval wing disc; B: third instar larval brain; WT: wild-type BEAF animal; KO: null BEAFAB-KO animal. The percent colocalization is comparable between WT and KO for both wing discs and brains, but is lower for the three negative controls.