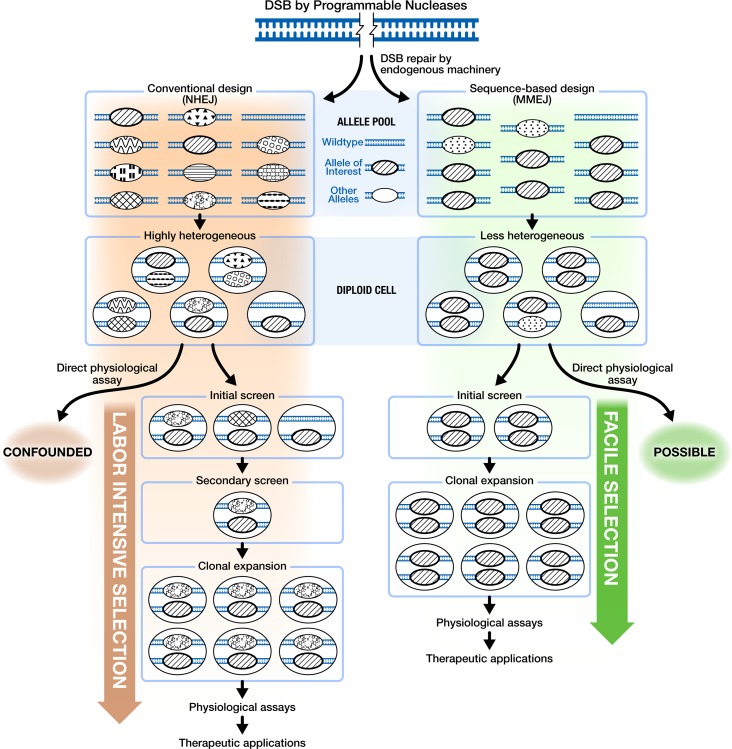
Fig 1. MMEJ is a unique DSB repair pathway that results in highly efficient and highly stereotyped mutagenesis.
DSB by conventionally designed Programmable Nucleases typically proceeds through a versatile yet unpredictable classical non-homologous end joining (NHEJ) pathway. As a result, a rather diverse cohort of mutant alleles are generated, making the subsequent selection process labor intensive to enrich for the allele of interest. The resulting genetic composition of the specific loci are often complex, requiring careful molecular characterization of each allele. Efficient activation of microhomology-mediated end joining (MMEJ) pathway, on the other hand, can greatly limit allelic diversity and enable the intentional generation of a particular deletion allele of interest at a high rate. Consequently, the downstream applications become more streamlined with facile generation of homozygous frameshift allele in diploid cells.