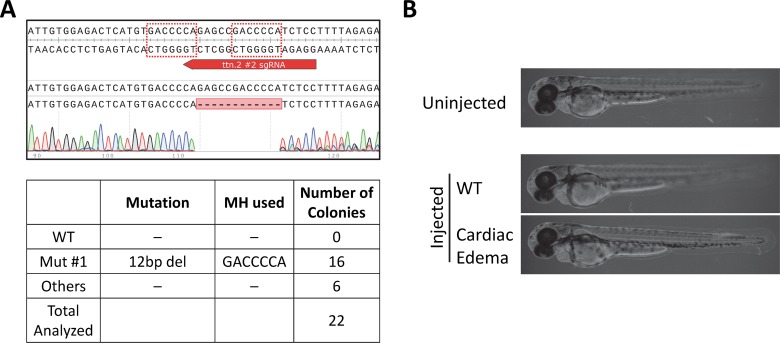
Fig 6. PreMA reagent can be used for in-frame gene alteration.
A. Top–Wildtype ttn.2 sequence with sgRNA target site annotated in red. The dotted red boxes are MH arms predicted to be used most frequently. Raw sequence alignment of the whole PCR amplicon demonstrates that the majority of reads are the expected 12 bp deletion allele. Bottom–summary data from subcloning analyses. 73% of the mutant allele recovered were of the predicted MH allele. B. 2 dpf zebrafish larvae injected with ttn.2 #2 sgRNA RNP (300 pg sgRNA + 660 pg Cas9) grossly appear normal with the exception of mild cardiac edema. Median penetrance was 50%. N = 3 biological and technical replicates. At least 9 injected animals were scored in each experiment.