Fig 7. Competition hypothesis version 2.
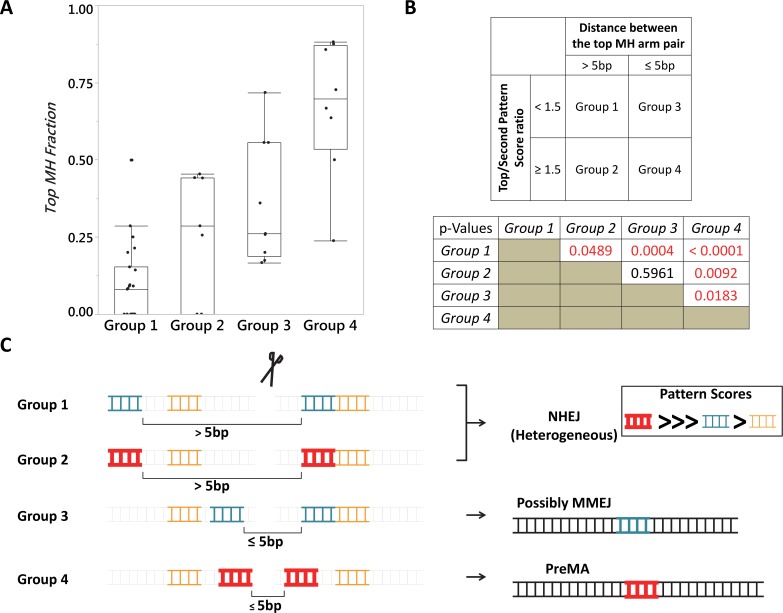
A. Outlier plot summarizing repair outcomes from 47 genomic targets using TALEN and CRISPR-Cas9. Close proximity of top predicted MH arms (Groups 3 and 4) appears to be the primary determinant for generating PreMA type outcomes as no target from Groups 1 and 2 had Top MH Fraction exceeding 0.5. When the top predicted allele had at least 50% higher Pattern Score than the second predicted allele (Groups 2 and 4), it was a strong indicator for inducing MMEJ-class repairs. B. Top Definition for each of the 4 groups used in Panel A. Each and every zebrafish genomic locus was segmented into these categories. Pattern scores were derived using RGEN online tool. Bottom P-values calculated by Wilcoxon’s Each Pair Calculation (adjusted for multiple comparisons). C. Graphical representation of each group detailed in Panel A. Groups 1 and 2 are prone to activate NHEJ-type outcomes, presumably because the yet-unidentified MMEJ factor fails to localize to suitable microhomology arm pairs, limited by how far apart these arms are. Group 4 is most suitable for strong MMEJ activation because it satisfies the proximity requirement AND the relative strength requirement. The latter may aid in the kinetics of the yet-unidentified MMEJ factor binding to the microhomology arms. Our data suggest that Group 3 is an intermediate group in terms of MMEJ activation. Perhaps extragenetic factors, such as cell cycle and epigenetic status may determine how favorable the loci are for MMEJ inductions.