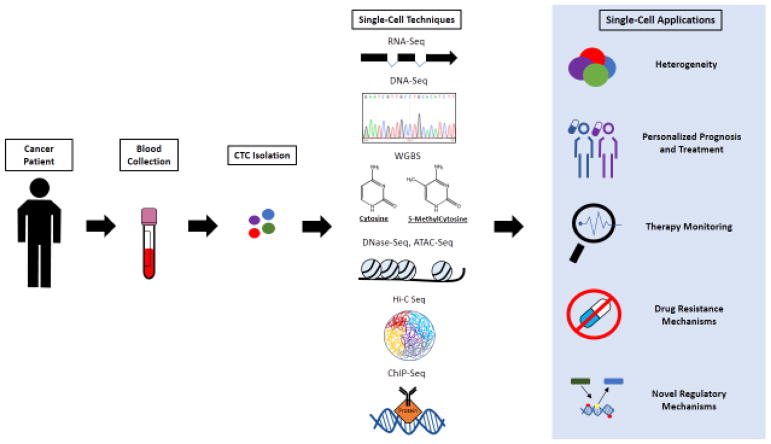
Figure 1, Key Figure. Illustration of current genome-wide technologies and potential applications for single CTCs.
Blood samples from cancer patients are processed and CTCs are isolated. DNA or RNA, or both, are isolated and processed by one or more of the following single-cell techniques: RNA-Seq, DNA-Seq, WGBS (whole genome bisulfite sequencing), DNase-Seq (DNase I hypersensitive Sequencing), ATAC-Seq (Assay for Transposase-Accessible Chromatin Sequencing), Hi-C Seq, ChIP-Seq (Chromatin Immunoprecipitation Sequencing). RNA-Seq depicts exomes and splicing probabilities. DNase-Seq and ATAC-Seq depicts nucleosomes on DNA. Hi-C Seq depicts an example of how DNA can be organized structurally within the nucleus. ChIP-Seq depicts a protein binding to DNA, which is captured by a targeting antibody. These techniques can manifest heterogeneous population of cells, personalize prognosis and treatment, enable therapy monitoring, manifest drug resistance mechanisms and lead to the discovery of novel regulatory mechanisms that may advance our understanding of cancer and make an impact on cancer treatments.