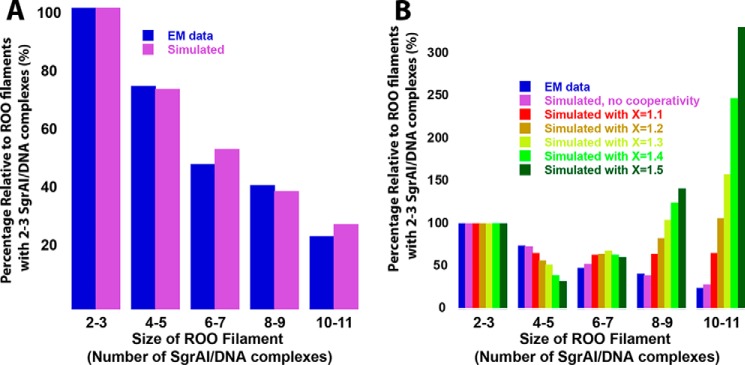
Figure 7.
Simulation of length or size distribution of run-on oligomers, with and without cooperativity. A, distribution normalized to the value for ROO filament sizes of 1–2. EM (blue) distribution data taken from the negative stain EM histogram (4), which used 3 μm SgrAI and PC DNA. Modeling (model EM) allowing up to 14 SgrAI–DNA complexes was used with rate constants from fitting Data Set 3 to model 4BM (Table 6) and initial concentrations of 3 μm SgrAI and PC DNA (shown in purple). B, as in A, but distribution of predicted ROO filament size with different degrees of cooperativity. Also shown is the distribution of oligomers (relative to the quantity of 2–3-mers) found using EM, in blue (4). To simulate positive cooperativity, the reverse rate constant for ROO filament dissociation was serially decreased by the given cooperativity factor (X = 1.1 to 1.5) with increasing ROO filament size, up to 5-mers, after which it remained constant (see “Experimental procedures”).