Abstract
We recently reported that PIK3CA mutant colorectal cancers (CRCs) are addicted to glutamine through up-regulation of glutamate pyruvate transaminase 2 (GPT2). A GPT2 inhibitor suppresses in vivo growth of PIK3CA mutant, but not wild-type, CRCs. This study indicates that targeting glutamine may be an effective approach to treat CRCs with PIK3CA mutations.
Metabolic reprogramming is a hallmark of cancer.1 It has long been known that most cancer cells are dependent on glutamine to grow. Although glutamine is a non-essential amino acid, it is a required supplement for culturing cancer cells. In addition to being used as a building block for protein, glutamine can also be utilized as a fuel source to replenish the tricarboxylic acid (TCA) cycle. In this process, glutamine is first converted to glutamate by glutaminases (GLSs), followed by conversion to α-ketoglutarate (α-KG), a TCA cycle intermediate (Fig. 1). The conversion from glutamate to α-KG is mediated by either transaminases or glutamate dehydrogenases. Recent studies showed that many oncogenes and tumor suppressor genes are involved in glutamine metabolism. PIK3CA, which encodes the p110α catalytic subunit of phosphatidylinositol 3-kinase α (PI3Kα), is the most frequently mutated oncogene in human cancers including 20–30% of colorectal cancers (CRCs).2, 3 However, whether mutant PIK3CA/p110α reprograms cancer metabolism was an important unaddressed question.
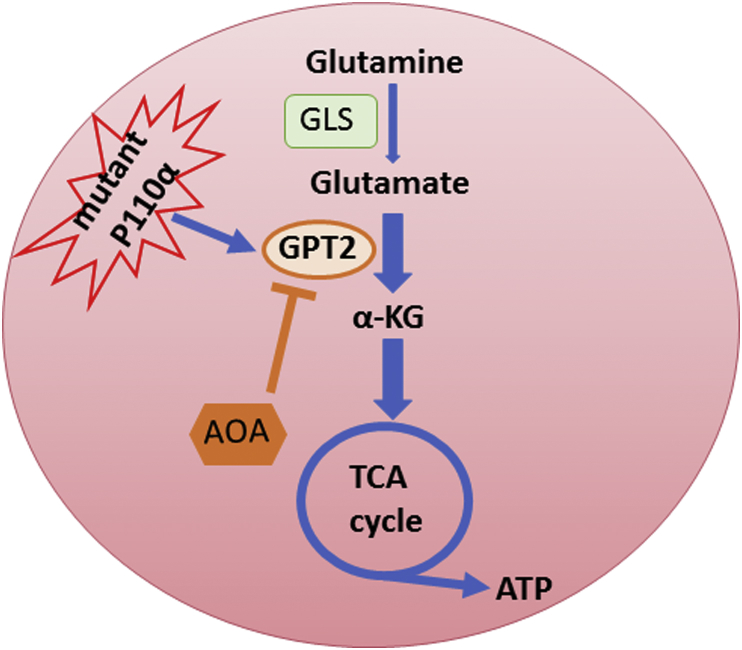
Fig. 1.
Oncogenic PIK3CA mutations reprogram glutamine metabolism by up-regulating GPT2. As a fuel source, glutamine is first converted to glutamate and then α-KG to replenish the tricarboxylic acid (TCA) cycle. Oncogenic p110α mutant protein reprograms glutamine metabolism through upregulation of GPT2. A pan-aminotransferase inhibitor AOA suppresses xenograft tumor growth of PIK3CA mutant, but not wild type (WT), CRCs. GLS: glutaminase; GPT2: glutamate pyruvate transaminase 2; α-KG: α-ketoglutarate; AOA: aminooxyacetate.
Using isogenic CRC cell lines with either the wild type (WT) or mutant alleles of PIK3CA knocked out, we demonstrated that PIK3CA mutations render CRCs more addicted to glutamine, but not glucose.4 To our knowledge, this is the first study to demonstrate that specific oncogenic mutations render cancer cells differentially sensitive to glutamine deprivation. Although it has been previously shown that WT KRAS regulates glutamine metabolism in pancreatic cancers,5 it is not clear that oncogenic KRAS mutations render cancer cells more sensitive to glutamine deprivation. Moreover, our study clearly demonstrated that KRAS mutations do not make colorectal cancers more dependent on glutamine, because isogenic KRAS mutant and WT CRC clones do not show differential sensitivity to glutamine deprivation.4
Mechanistically, mutant PIK3CA/p110α up-regulates mitochondrial glutamate pyruvate transaminase 2 (GPT2) and therefore converts more glutamate to α-KG to replenish the TCA cycle,4 which generates more ATP and macromolecules to sustain rapid tumor growth. Although AKT is a well-known downstream mediator of the PI3K signaling, mutant p110α up-regulates GPT2 gene expression by an AKT independent pathway. Mutant p110α activates RSK2 kinase through PDK1.4 Activated RSK2 then phosphorylates ATF4 at the serine residue 245, which in turn recruits the deubiquitinase USP8 and protects ATF4 from ubiquitin-mediated degradation.4 Accumulation of ATF4 enhances GPT2 gene expression. Therefore, our study revealed a novel p110α–PDK1–RSK2–ATF4–GPT2 signaling axis that reprograms glutamine metabolism.
Our study has important therapeutic implications. Despite huge efforts made by both pharmaceutical companies and academic institutions in last ten years, only a couple of p110α-specific inhibitors have been developed. Early clinical trials of the Novartis p110α-specific inhibitor, BYL719, indicate that BYL719 alone induced partial responses in cancer patients harboring PIK3CA mutations.6 Recent studies showed that cancers quickly develop resistance to BYL719 by loss of PTEN or up-regulation of AXL.7 Therefore, alternative approaches are urgently needed to target PIK3CA mutations in patients. We demonstrated that aminooxyacetate (AOA), a compound that targets glutamine metabolism by blocking conversion of glutamate to α-KG (Fig. 1), effectively inhibits xenograft tumor growth of four colorectal cancer cell lines harboring PIK3CA mutations.4 In contrast, AOA has no effect on the growth of xenograft tumors established from two colorectal cancer cell lines with WT PIK3CA.4 Although AOA is a pan-aminotransferase inhibitor, our data clearly demonstrate that the tumor inhibitory effect of AOA on PIK3CA mutant xenografts is indeed on-target to GPT2, because GPT2 knockdown cells grow much slower in nude nice and these tumors are insensitive to AOA treatment.4 It is conceivable that a GPT2-specific inhibitor could be more potent and less toxic. Our current effort is devoted to develop potent and specific GPT2 inhibitor by performing a high throughput compound screening campaign and chemically modifying the lead compound AOA.
Lastly, our study suggest that PIK3CA mutations may serve as a predictive biomarker for drugs that target glutamine metabolism. Predictive markers are critical tools to guide successful clinical trials and effective treatment of cancer patients. For example, in lung cancer, only patients with EGFR mutations respond to EGFR-inhibitor treatment, whereas in colon cancers acquired KRAS mutations predict resistance to anti-EGFR antibody therapies. These markers provide valuable tools for guiding the choice of cancer therapies. Increasing evidence indicates that targeting glutamine metabolism could be an effective cancer therapy. Many preclinical studies demonstrated that targeting glutamine metabolism effectively inhibits tumor growth of a variety of human cancers.8 Several compounds that target glutamine metabolism pathways are under development. In fact, phase I clinical trials have been undertaken for a compound (CB-839) that inhibits glutaminase.9 A key question is how to better select cancer patients to be treated with drugs that target glutamine metabolism. Our unpublished data demonstrated that CB-839 also inhibits xenograft tumor growth of PIK3CA mutant CRCs, but not PIK3CA WT CRCs, suggesting that PIK3CA mutations may serve as a biomarker to predict patients who respond to drugs targeting glutamine metabolism.
In summary, implications of our study are three-fold: (1) shed new lights into the mechanisms by which PIK3CA mutations drive colorectal tumorigenesis through reprogramming glutamine metabolism; (2) identify GPT2 as a drug target for CRCs with PIK3CA mutations; and (3) indicate PIK3CA mutation as a predictive biomarker for drugs targeting glutamine metabolism. Future studies are warranted to pursue these exciting directions.
Acknowledgement
This work is supported by National Institutes of Health grants R01CA196643, R01CA127590, P50CA150964 and P30 CA043703.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Samuels Y., Wang Z., Bardelli A. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 3.Lawrence M.S., Stojanov P., Mermel C.H. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao Y., Samuels Y., Li Q. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat Commun. 2016;7:11971. doi: 10.1038/ncomms11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Son J., Lyssiotis C.A., Ying H. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juric D., Castel P., Griffith M. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kalpha inhibitor. Nature. 2015;518:240–244. doi: 10.1038/nature13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elkabets M., Pazarentzos E., Juric D. AXL mediates resistance to PI3Kalpha inhibition by activating the EGFR/PKC/mTOR axis in head and neck and esophageal squamous cell carcinomas. Cancer Cell. 2015;27:533–546. doi: 10.1016/j.ccell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hensley C.T., Wasti A.T., DeBerardinis R.J. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross M.I., Demo S.D., Dennison J.B. Antitumor Activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014;13:890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]