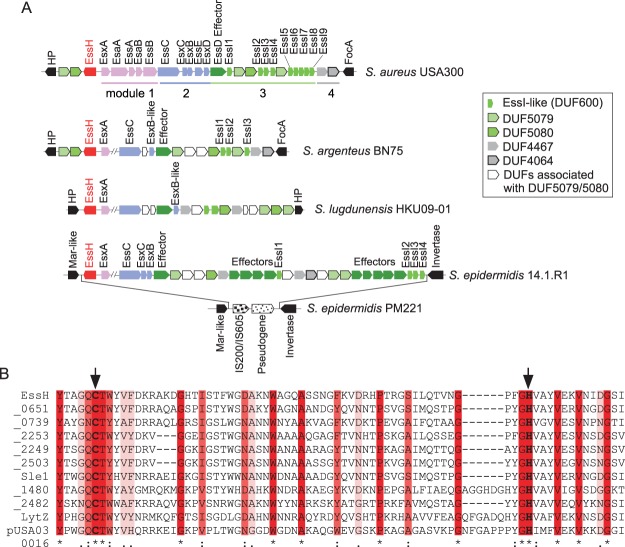
FIG 1.
Association of essH with the Staphylococcus aureus ESS cluster. (A) Genomic organization of ESS clusters of S. aureus USA300 LAC, Staphylococcus argenteus BN75, Staphylococcus lugdunensis HKU09-01, Staphylococcus epidermidis 14.1.R1, and S. epidermidis PM221. Genes in modules 1, 2, 3, and 4 are shown in pink, blue, shades of green, and gray, respectively. Genes flanking the cluster are shown in black; essH is shown in red. DUFs (domains of unknown function) encoded by module 4 genes are listed in the inset. In some strains, module 4 genes are also found upstream of essH. The complete set of modules 1 and 2 genes is only shown for S. aureus USA300 LAC; these genes are conserved in the other species. FocA, formate channel A; HP, hypothetical protein; MarR, multiple antibiotic resistance regulator. (B) CHAP domains of 11 proteins predicted from the genome sequence of S. aureus subsp. aureus USA300_FPR3757 (NCBI RefSeq accession number NC_007793.1) were aligned with Clustal W. The locus tag for essH is SAUSA300_0277. Proteins are aligned with EssH in order of declining sequence conservation. Genes whose products encompass CHAP domains are identified with the last four digits of their locus tag; SAUSA300_0438 and _2579 are listed as Sle1 and LytZ. TraG (pUSA300016) is encoded on a plasmid. Identical, conserved, and similar residues are denoted by asterisks, colons, and periods, respectively, and highlighted in shades of red. Arrows identify the predicted active site residues, Cys199 and His254.