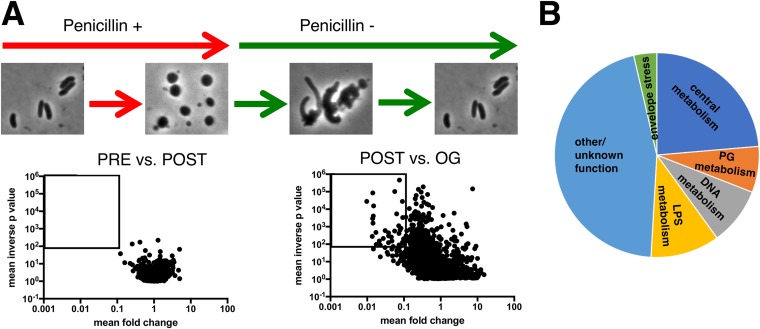
FIG 5.
Identification of genes required for sphere recovery with TnSeq. (A) Schematic of experimental design and volcano plots of change in relative abundance of insertion mutants between the two conditions (x axis) versus the concordance of independent insertion mutants within each gene (y axis, inverse P value). The square denotes the cutoff criteria applied (>10-fold fitness defect, P < 0.01) for identification of genes contributing to sphere recovery. PRE versus POST is a comparison of insertion frequencies after/before antibiotic exposure, and POST versus OG is a comparison between an outgrowth period and directly after antibiotic exposure (see Materials and Methods for details). (B) Distribution of the main functional categories of gene insertions that confer a postantibiotic fitness defect. Functional categories were assigned manually following annotation of individual proteins in the Kegg database (http://www.genome.jp/kegg/).