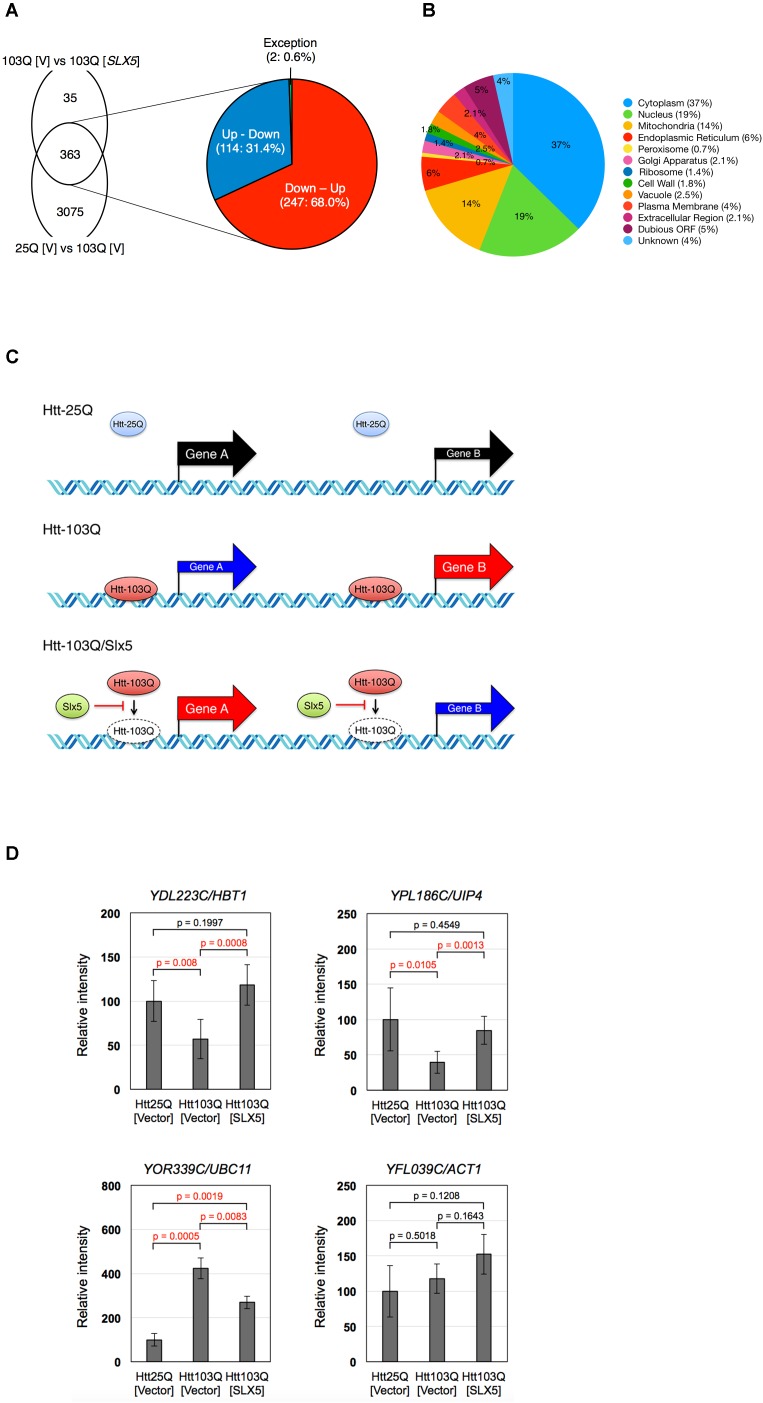
FIGURE 5.
Slx5 modulates the transcriptional activity due to expression of Htt-103Q-NLS. Depiction of 398 SLX5 modulated genes identified by RNA sequencing. (A) RNA-seq analysis shows that Htt103Q-NLS leads to a global effect on the transcriptome as it affects the expression of 3438 genes (25Q [V] vs. 103Q [V]). Plasmid-borne SLX5 affects the expression of 398 genes in Htt103Q-NLS cells (103Q [V] vs. 103Q [SLX5]) as shown in Supplementary Table S2. The overlap between Htt25Q-NLS and Htt103Q-NLS (25Q [V] vs. 103Q [V]) is 363 genes. The expanded view of A shows that expression of most of the 363 genes (99.4%) inversely correlates between 25Q [V] vs. 103Q [V] and 103Q [V] vs. 103Q [SLX5]. Expression of 247 of the 363 genes (68.0%) is down-regulated by Htt103Q, and this effect is reversed by plasmid-borne SLX5 (Down – Up). Expression of 114 of the 363 genes (31.4%) is up-regulated by the Htt103Q, and this effect is reversed by plasmid-borne SLX5 (Up – Down). (B) Subcellular localization of differentially expressed genes. The localization of proteins encoded by the 398 genes indicated in Supplementary Table S2 was analyzed using cellular components assignment from the PANTHER Classification System and the Saccharomyces Genome Database. Pie chart shows a ratio of the genes placed into cellular component categories. Individual genes are listed in Supplementary Table S4. (C) Schematic of gene expression in Htt25Q-NLS and Htt103Q-NLS cells and the effect of plasmid-borne SLX5 on the transcriptome of Htt103Q-NLS cells. Expression of genes A and B are downregulated and upregulated in Htt103Q-NLS cells relative to Htt25Q-NLS cells, respectively. Slx5 reduces the association of Htt with chromatin and this contributes to the reversal in gene expression such that gene A is upregulated and gene B is downregulated. (D) RT-PCR validation of gene expression analysis. Total RNAs were purified from strains expressing either Htt25Q-NLS or Htt103Q-NLS from a GAL promoter for 4 h with or without plasmid-borne SLX5. RT-PCR analyses was performed using the same samples used for the RNA-seq. Relative intensities are reported as the mean ±SD of three biological repeats. Reactions for YDL223C/HBT1 and YPL186C/UIP4 were performed in duplicate. N = 3 for YOR339C/UBC11 and YFL039C/ACT1, N = 6 for YDL223C/HBT1 and YPL186C/UIP4.