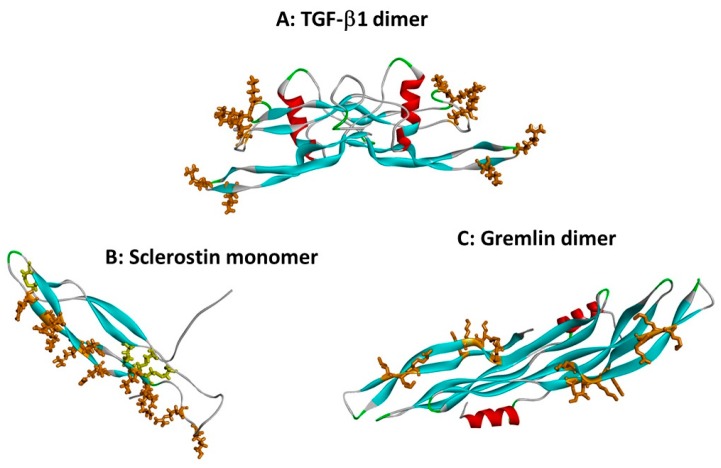
Figure 2.
Heparin binding sites on TGF-β superfamily cytokines. Protein chains are shown as in Figure 1, and heparin binding site basic residues are shown in brown stick format. (A) The dimer of TGF-β1 (co-ordinates from 1KLC.pdb). Residues K25, R26, K31, K37, R94 and R97 form a discontinuous heparin binding site at the tips of the “fingers” [13]. (B) Sclerostin monomer (one of the NMR ensemble in 2K8P.pdb) with heparin-binding residues in brown. Cystine residues are shown in yellow stick format. Loop 3 (the second β-strand loop) and Loop 2 are both involved; residues K99, R102, R114, R116, R119, R131, R133, K134, R136, K142, K144 and R145 form a linear heparin binding site capable of accommodating a heparin dodecamer [26]. (C) The dimer of the CAN BMP antagonist gremlin (co-ordinates from 5AEJ.pdb). Here, the heparin binding residues are located largely along the second “finger”, as for sclerostin; in the dimer both copies of the heparin binding site are on the concave face. Residues K145, K147, K148, K167, K168, K169, K174 and R177 have been identified as forming the heparin binding site [20]. The mode of dimerisation and the location of the heparin binding site both differ from those of TGF-β1.