Figure.
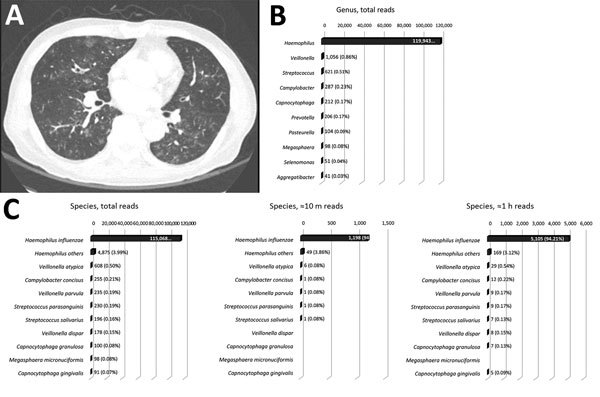
Chest computed tomography scan and sequencing of the 16S amplicon in a 77-year-old man with end-stage renal disease and asthma. A) Hypoxic respiratory failure with bilateral infiltrates are visible on chest computed tomography scan. B) Sequencing of the 16S amplicon performed on sputum using the MinION sequencer (Oxford Nanopore Technologies, Oxford, UK). Sequencing for 5 h generated 470,231 reads. A total of 122,272 reads were aligned with 1 of the bacterial 16S rRNA gene sequences, and most reads (119,943 [98.1%]) were aligned with genus Haemophilus. C) Of the 122,272 aligned reads, nearly all (115,068 [94.11%]) were aligned with the species H. influenzae (left). The number of reads aligned with H. influenzae was >100-fold larger than those aligned with other oral commensal bacteria. Similar results were obtained from the subgroup analyses of reads generated during the first hour (middle) and during the first 10 min (right).
