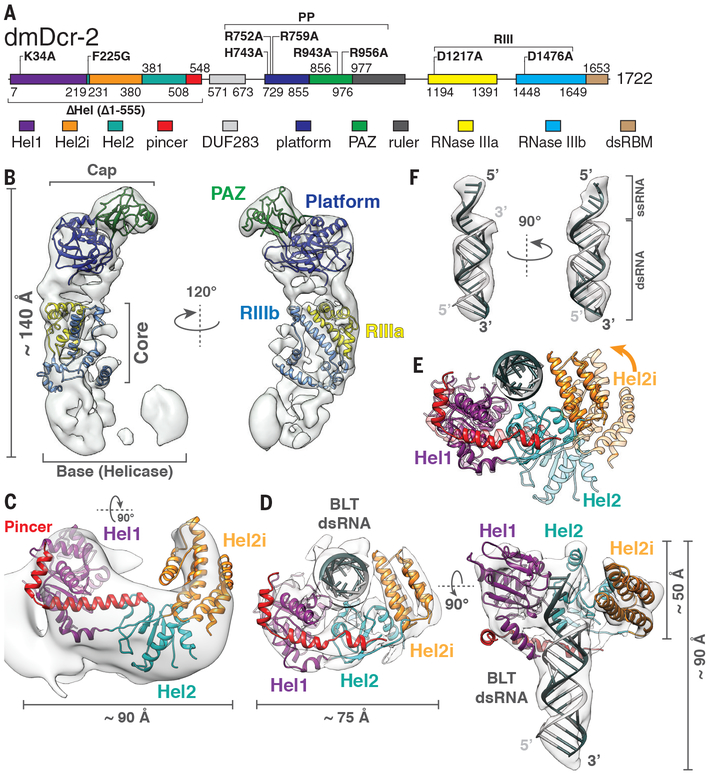
Fig. 1. Cryo-EM reconstructions of apo-dmDcr-2 and dmDcr-2•BLT dsRNA•ATP-γS.
(A) dmDcr-2 domains numbered at boundaries. Mutations/deletions in bold, and designated in text as superscripts. All mutations were in the context of the full-length protein, unless specified by Δ. Two full-length variants had multiple mutations: PP (H743A, R752A, R759A, R943A, R956A); RIII (D1217A, D1476A) (B) Cryo-EM density map of apo-dmDcr-2 (7.1 Å) fitted with homology models of subdomains. RIIIa, RNase IIIa; RIIIb, RNase IIIb. (C) Homology model of apo-dmDcr-2 in open conformation, based on apo-RIG-I, and fitted as rigid body into 8.7 Å cryo-EM density map (also Fig. S4A and Fig. S5, B to D). (D) Cryo-EM reconstruction of dmDcr-2RIII•BLT dsRNA•ATP-γS showing helicase in closed, ligand-bound conformation. (E) Superimposition of open (light) and closed (dark) helicase conformations showing clamping of Hel2 and Hel2i on BLT dsRNA. Arrow, direction of clamping. (F) EM density and modeling of BLT single and double-stranded RNA.