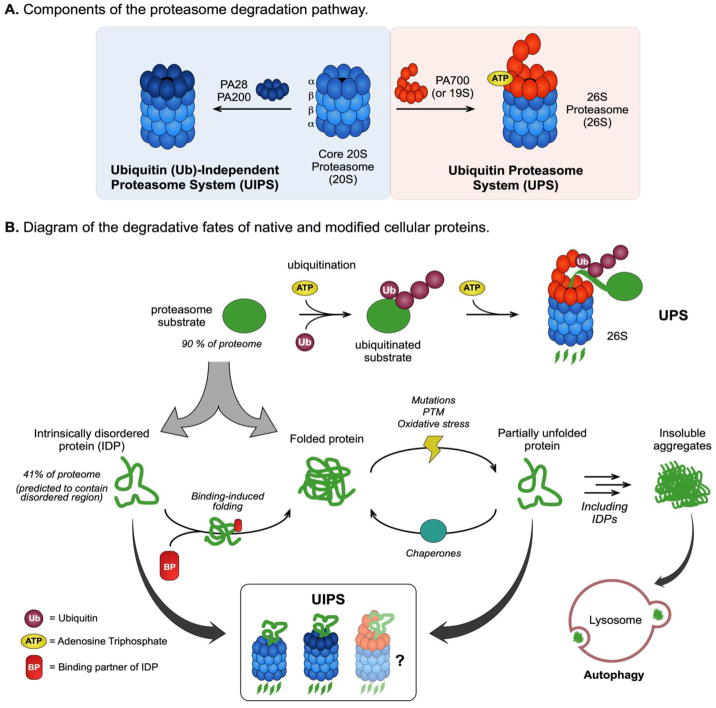
Figure 1. Structure, Function, and Substrate Profile of the Proteasome Degradation Pathway.
(A) Components of the ubiquitin proteasome system (UPS) and the ubiquitin (Ub)-independent proteasome system (UIPS). (B) Diagram of substrate turnover by the different proteasome pathways. The 26S proteasome degrades over 90% of the proteome, including intrinsically disordered proteins (IDPs) and folded proteins, by the Ub- and ATP-dependent UPS. The UIPS targets substrates independent of Ub-conjugation and can effectively degrade IDPs, which constitute up to 41% of the proteome, but not folded proteins due to their three-dimensional structure. Cellular stresses, including mutations, post-translational modifications (PTMs) and oxidative damage, can partially unfold structured proteins making them susceptibility to turnover by the UIPS. Partially unfolded proteins (and IDPs) can self-associate to form insoluble aggregates, which cannot be processed by the proteasome, and are predominantly cleared from the cell through the lysosome-autophagy pathway.