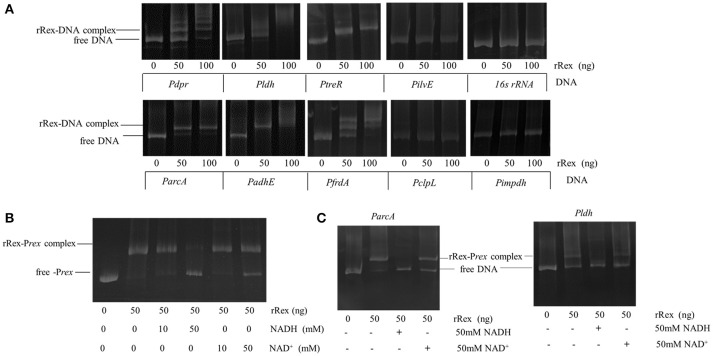
Figure 6.
Determination of Rex binding abilities by EMSAs. (A) Binding of purified rRex to the promoter regions of different genes using EMSAs. The resulting protein–DNA complexes were separated from unbound DNA fragments using native polyacrylamide gels. The DNA fragments were visualized by ethidium bromide staining. Formation of stable Rex–DNA complexes resulted in one or more distinct shifted DNA bands. The promoter region of impdh, which is not regulated by Rex and whose promoter sequence lacks a putative Rex binding box, and 16sRNA DNA fragments were used as negative controls. (B) EMSAs were performed with PCR products of the promoter regions of rex incubated with 50 ng purified Rex protein and different concentrations of NAD+ and NADH. At lower concentration, NADH and NAD+ did not influence the rRex affinity to Prex and not change the mobility pattern. A higher concentration of NADH (50 mM) completely prevents the formation of rRex–Prex complexes, and 50 mM of NAD+ causes the complexes to dissociate slightly. (C) EMSAs were performed with PCR products of the promoter regions of arcA and ldh incubated with 50 ng purified rRex protein and 50 mM NAD+ or 50 mM NADH. The presence of 50 mM NADH inhibits promoter-DNA complex formation for ParcA and Pldh. However, the binding affinity between rRex and ParcA is slightly decreased in the presence of 50 mM NAD+, and rRex binds better to the Pldh when compared to ParcA under the same conditions.