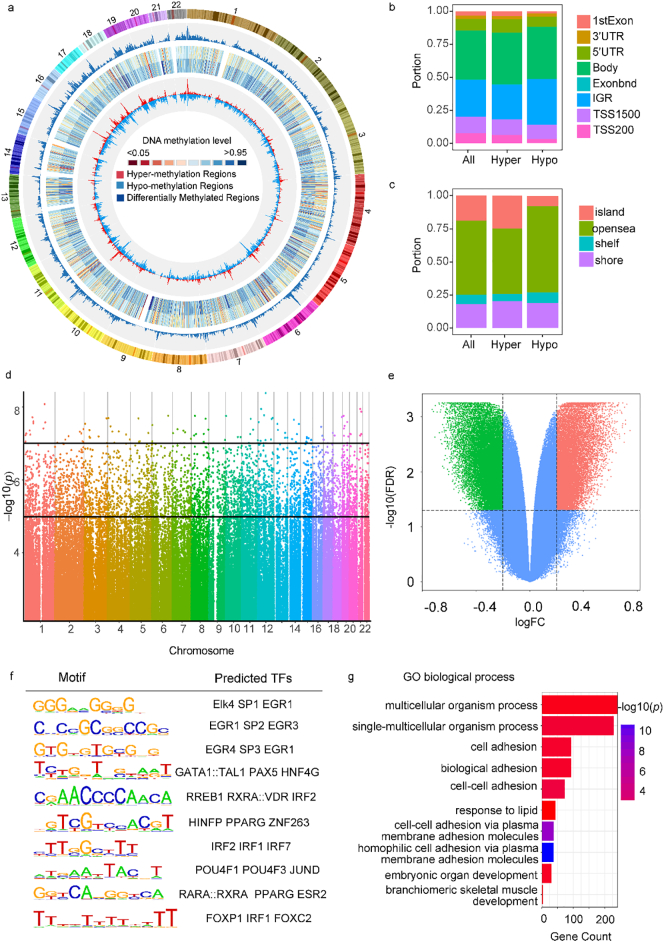
Fig. 1.
DNA methylation patterns in RPL and controls. (a) Circular representation of whole genome DNA methylation level for RPL and controls. CpG methylation level were averaged per Mb in the genome and represent as histograms and heatmaps. (b) Distributions of hyper-methylated and hypo-methylated DMPs were investigated throughout each genomic region, FDR < 0·05. (c) Genomic locations divided into island, opensea, shelf and shore according to the distance between CpG island and DMPs, FDR < 0·05. (d) Manhattan plot showing P value based on DMPs analyses. The two horizontal lines is the suggestive DNA methylation chip significance threshold cutoff. (e) A volcano plot of the distribution between FDR (adjusted P value) and difference in β value (logFC). Lines represent used cutoff values to identify the most hyper-methylated DMPs (red) and hypo-methylated DMPs (green). (f) DNA sequences motifs identified to be enriched in DMRs by CisGenome Browser software. The right column contains the top three transcription factor similar to these motifs using JASPAR database. (g) Gene enrichment analysis of gens with DMPs in RPL.