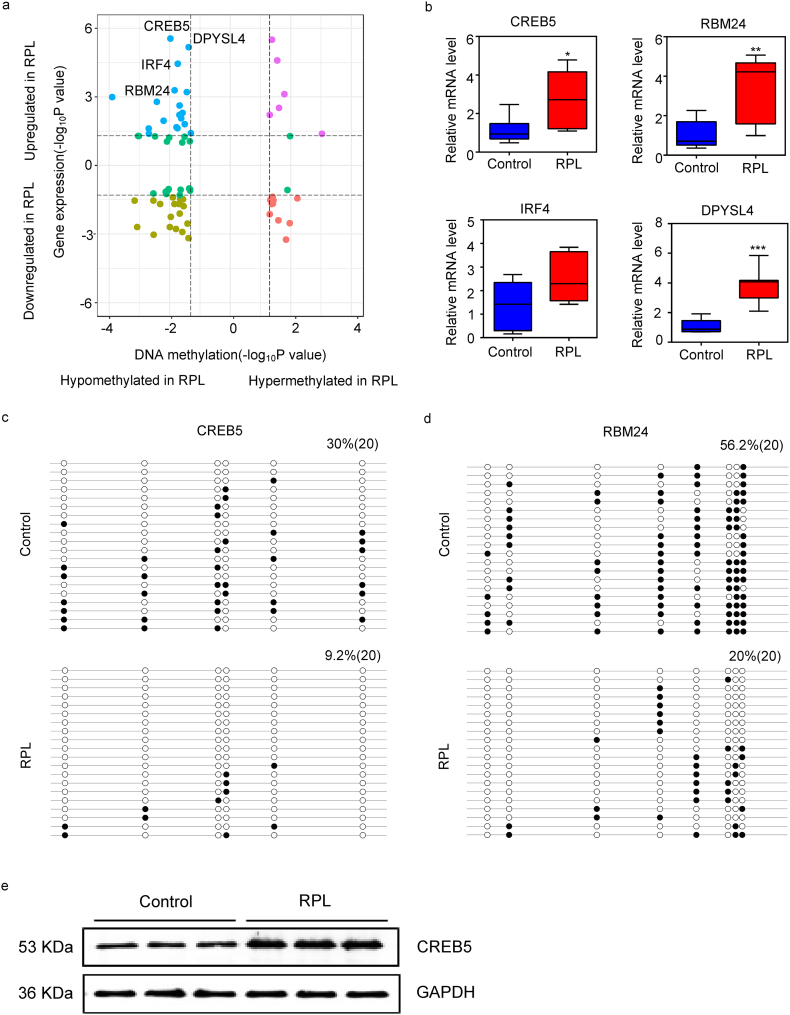
Fig. 2.
Filtration of genes with synergetic alterations in DNA methylation and gene expression. (a) Quadrant plot showing DMRs and expression of corresponding genes. The dashed lines indicate a threshold of P value below 0·05. The blue dots signify hypo-methylated and up-regulated genes in RPL, the brown bots signify hypo-methylated and down-regulated genes in RPL, the purple bots signify hyper-methylated and up-regulated genes in RPL and the red bots signify hyper-methylated and down-regulated genes in RPL. (b) Box plots of relative expression level of four genes selected by a threshold of 5% FDR. Error bars, SEM. *P < 0·05, **P < 0·01, ***P < 0·001. Bisulfite sequencing of CREB5 (c) and RBM24 (d). Open and closed circles represent unmethylated and methylated CpG sites and each row corresponds to a single clone sequenced. All loci shown in picture were found to be hypo-methylated in RPL according to the analysis of Infinium Human Methylation 850 K BeadChip data. (e) The expression of CREB5 protein in control group and RPL group.