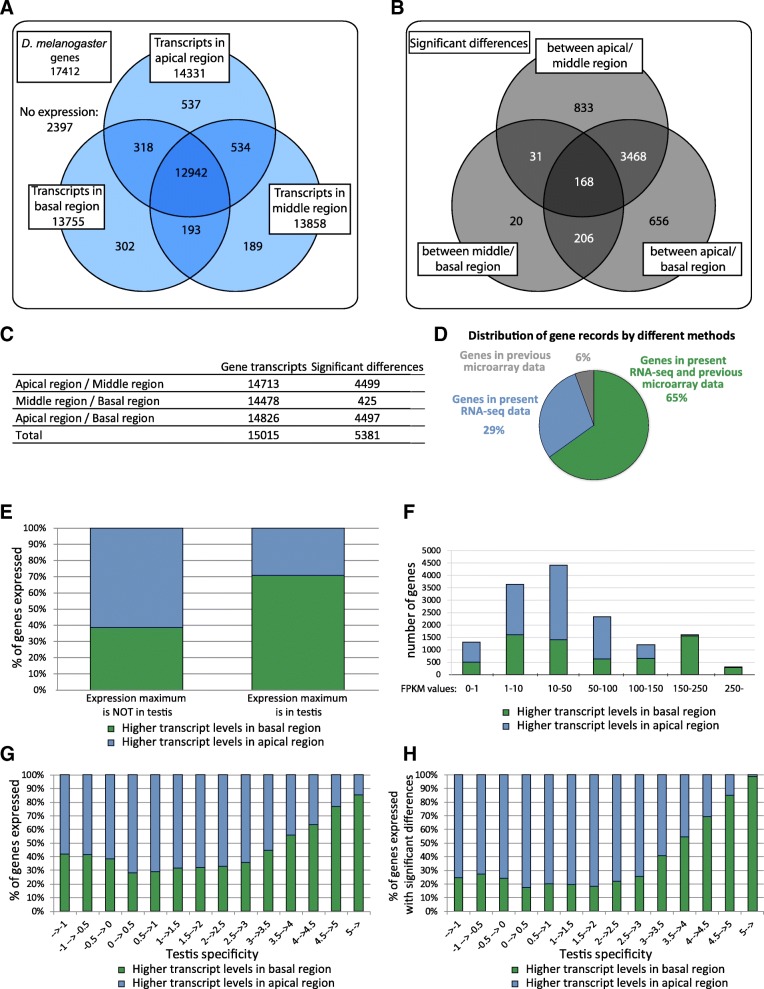
Fig. 1.
Distribution of transcripts in different region of testis and analysis of the RNA-Seq results. a Distribution of Drosophila melanogaster transcripts by RNA-Seq. b Number of genes showing significant differences (FDR-corrected p-value< 0.05) in transcript level between different region of testis. c The combined number of transcripts and the significant differences (FDR-corrected p-value< 0.05) between different region of testis. d Comparison of the previous microarray data [7] with present RNA-seq data. e Distribution of genes with lower or higher transcript levels in the basal testicular regions compared to the apical region. The comparison was based on the tissue where their expression maximum is defined by modENCODE database [56]. f Distribution of transcript level differences in basal region compared to the apical region as a function of gene expression levels. g Distribution of differences in transcript levels along the apical-basal axis of the testis as a function of tissue specificity index. h Distribution of genes with statistically significant transcript levels along the apical-basal axis of the testis as a function of tissue specificity index