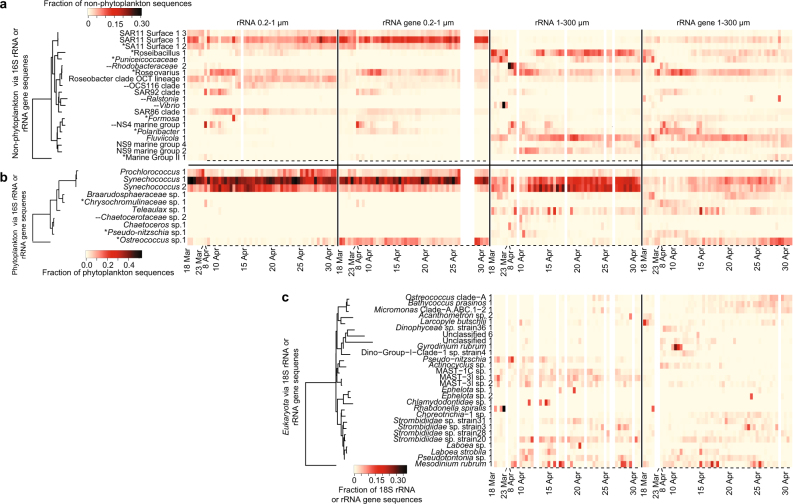
Fig. 3.
Daily to semi-daily 16S and 18S rRNA and rRNA gene dynamics of microbial taxa. Heatmaps include data from (a) “non-phytoplankton” Bacteria and Archaea via 16S rRNA and rRNA gene sequences, (b) “phytoplankton”, via 16S rRNA and rRNA gene sequences of chloroplasts and Cyanobacteria, and (c) Eukaryota taxa via 18S rRNA and rRNA gene, excluding metazoan sequences. Only ASVs or OTUs that ever became the taxon with the highest proportion of sequences within a given data set for at least one sample are shown. The tree shows the phylogenetic relatedness of the ASV or OTU according to the amplicon-sequenced region. Note that Mesodinium is known to have a very aberrant 18S rRNA gene sequence [49]. For the dates where two samples were taken per day (10:00 a.m. and 10:00 p.m., 10 April–1 May), a dash underneath a given sample indicates the sample was taken at night. All 16S rRNA and rRNA gene ASVs shown here were also detected during the 2011 diatom bloom study [7, 10], except where “--” is found next to the ASV name; asterisks next to taxon names indicate that ASV was also found to most abundant during the 2011 study.