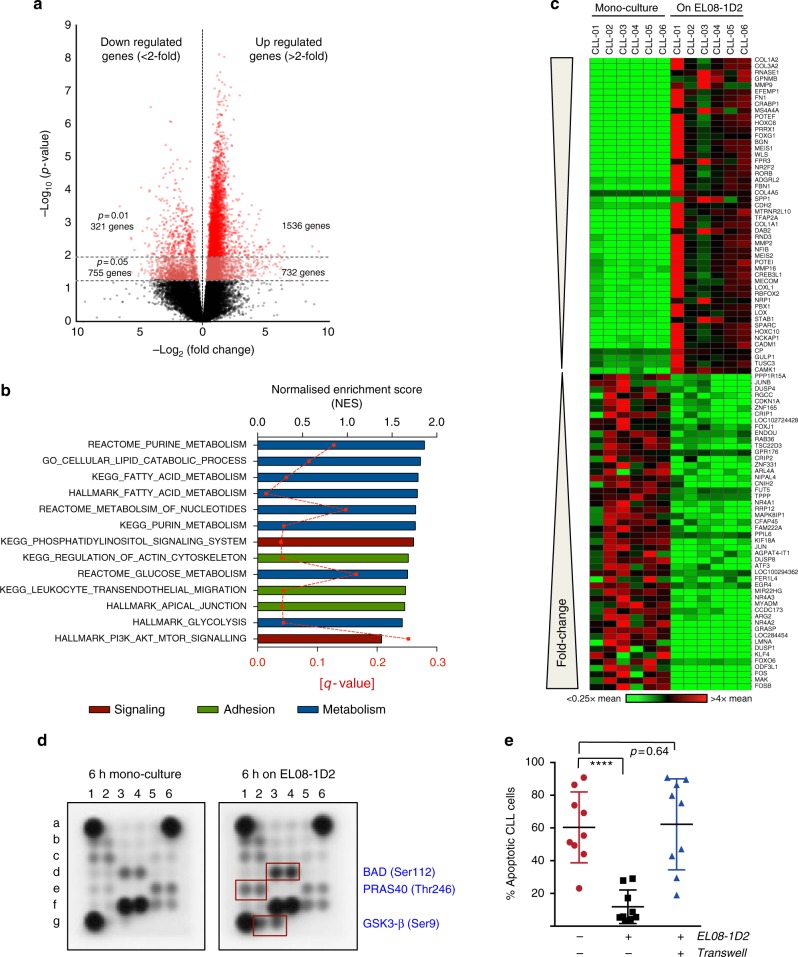
Fig. 1.
Activation of CLL cells by BSMCs. a Volcano plot showing the differentially up- and down-regulated genes in CLL cells after 48 h of co-culture on EL08-1D2 cells compared to cells cultured for 4 h in mono-culture (4 h was chosen to avoid gene expression changes related to cell death). RNA-sequencing was performed on samples from 6 individual patients. b Transcriptomic data were subjected to Gene Set Enrichment (GSEA) analyses to identify pathways in CLL activated by contact to stromal cells. Gene sets are listed in order of Normalised Enrichment Scores (top black X-axis). FDR q values for each gene set are indicated by the red dotted line (lower red X-axis). c Heat map showing the 50 most significantly up- and down-regulated genes in CLL cells in response to contact with stromal cells. d Cell extracts from CLL mono-culture or from cells cultured for 6 h on EL08-1D2 cells were analysed using a human intracellular phosphorylation antibody array. Representative results from four different patients and experiments are shown. e CLL cells were cultured in medium only (red circles) or on EL08-1D2 cells (black squares) for 5 days before analysing apoptotic cells by Annexin-V/PI staining. Transwells were used to disrupt direct cell–cell contacts (blue triangles). Error bars show mean ± SEM from 9 patients; ****p < 0.0001