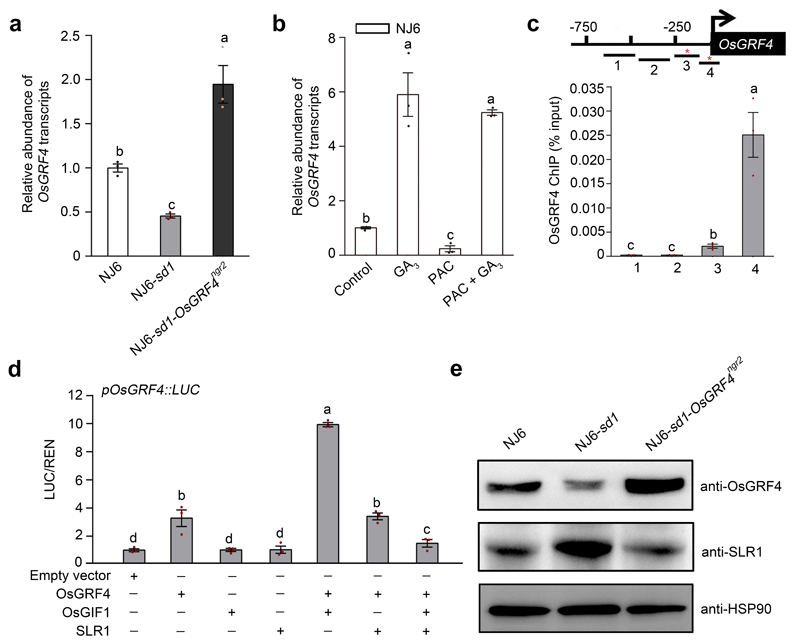
Extended Data Figure 6. SLR1 inhibits OsGRF4-OsGIF1 self-promotion of OsGRF4 mRNA and OsGRF4 protein abundance.
a, OsGFR4 mRNA abundance, plant genotypes as indicated. Abundance shown relative to that in NJ6 (=1). Data shown as mean ± s.e.m. (n = 3). Different letters denote significant differences (P < 0.05, Duncan's multiple range test). b, The effects of GA and PAC on OsGRF4 mRNA abundance in 2-week-old NJ6 plants. Abundance shown relative to that in water treatment control (=1). Data shown as mean ± s.e.m. (n = 3). Different letters denote significant differences (P < 0.05, Duncan's multiple range test). c, ChIP-PCR OsGRF4-mediated enrichment (relative to input) of GCGG-containing OsGRF4 promoter fragments (marked with *). Data shown as mean ± s.e.m. (n = 3). Different letters denote significant differences (P < 0.05, Duncan's multiple range test). d, OsGRF4-activated promotion of transcription from the OsGRF4 gene promoter-luciferase reporter construct is enhanced by OsGIF1 and inhibited by SLR1. Abundance of LUC/REN shown relative to that in empty vector control (=1). Data shown as mean ± s.e.m. (n = 3). Different letters denote significant differences (P < 0.05, Duncan's multiple range test). e, OsGRF4 abundance (as detected by an anti-OsGRF4 antibody), plant genotypes as indicated. HSP90 serves as loading control. The pictures of western blots represent one of the three experiments performed independently with similar results.