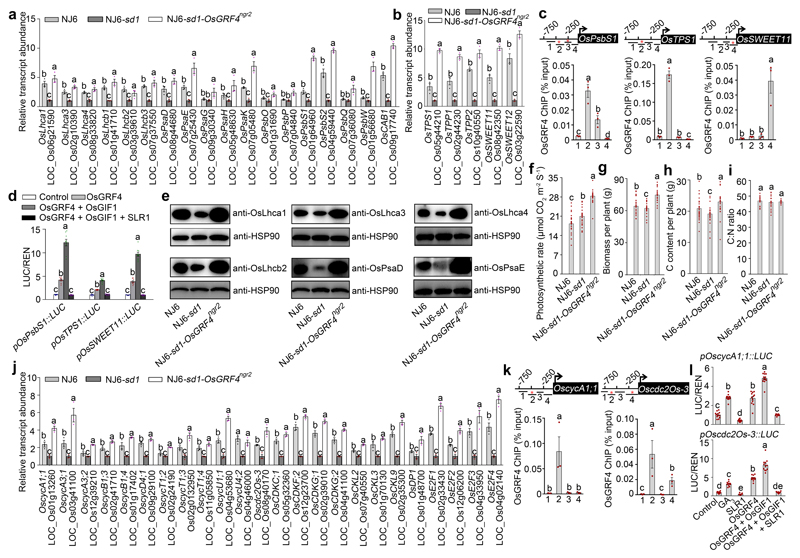
Extended Data Figure 7. The OsGRF4-SLR1 antagonism regulates carbon assimilation and plant growth.
a, b, Relative shoot abundances of C-fixation gene mRNAs. Abundances of transcripts of genes regulating photosynthesis (a), sucrose metabolism and transport/phloem loading (b) in NJ6, NJ6-sd1 and NJ6-sd1-OsGRF4ngr2 plants. Data shown as mean ± s.e.m. (n = 3). Abundances in NJ6 and NJ6-sd1-OsGRF4ngr2 expressed relative to NJ6-sd1 (= 1). Different letters denote significant differences (P < 0.05, Duncan's multiple range test). c, ChIP-PCR assays. Diagram depicts the OsPsbS1, OsTPS1 and OsSWEET11 promoters and regions used for ChIP-PCR, and GCGG-containing promoter fragment (marked with *) enrichment (relative to input). Data shown as mean ± s.e.m. (n = 3). Different letters denote significant differences (P < 0.05, Duncan's multiple range test). d, Transactivation assays. The LUC/REN activity obtained from a co-transfection with an empty effector construct and indicated reporter constructs was set to be one. Data shown as mean ± s.e.m. (n = 9). Different letters denote significant differences (P < 0.05, Duncan's multiple range test). e, Immunoblot detection of OsLhca1, OsLhca3, OsLhca4, OsLhcb2, OsPsaD and OsPsaE using antibodies as shown in genotypes as indicated. HSP90 serves as loading control. The pictures of western blots represent one of the three experiments performed independently with similar results. f-i, Comparisons of photosynthetic rates (f), biomass (g), C content (h) and C:N ratio (i) among NJ6, NJ6-sd1 and NJ6-sd1-OsGRF4ngr2 plants. Data shown as mean ± s.e.m. (n = 30). Different letters denote significant differences (P < 0.05, Duncan's multiple range test). j, Relative shoot abundances of mRNAs transcribed from cell-cycle regulatory genes in NJ6, NJ6-sd1 and NJ6-sd1-OsGFR4ngr2 plants. Transcription relative to the level in NJ6-sd1 plants (set to one). Data shown as mean ± s.e.m. (n = 3). Different letters denote significant differences (P < 0.05, Duncan's multiple range test). k, ChIP-PCR assays. Diagram depicts the OscycA1.1 and Oscdc2Os-3 promoters and regions (GCGG-containing fragment marked with *) used for ChIP-PCR. Data shown as mean ± s.e.m. (n = 3). Different letters denote significant differences (P < 0.05, Duncan's multiple range test).l, Transactivation assays from the OscycA1.1 and Oscdc2Os-3 promoters. Data shown as mean ± s.e.m. (n = 12). Different letters denote significant differences (P < 0.05, Duncan's multiple range test).