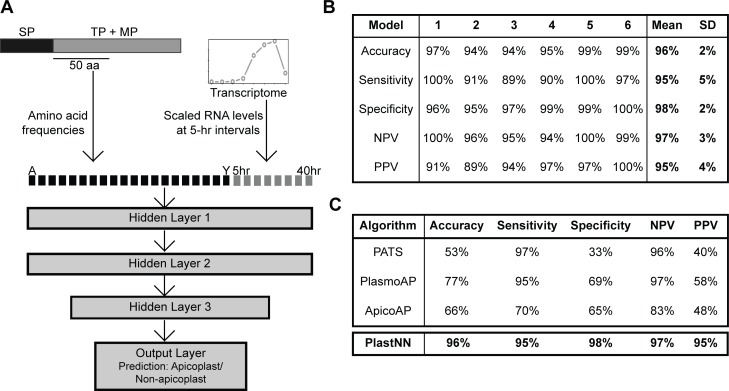
Fig 4. Improved prediction of apicoplast proteins using the PlastNN algorithm.
(A) Schematic of the PlastNN algorithm. For each signal peptide-containing protein, a region of 50 amino acids immediately following the signal peptide cleavage site was selected, and the frequencies of the 20 canonical amino acids in this region were calculated, resulting in a vector of length 20. Scaled RNA levels of the gene encoding the protein at 8 time points were added, resulting in a 28-dimensional vector representing each protein. This was used as input to train a neural network with 3 hidden layers, resulting in a prediction of whether the protein is targeted to the apicoplast or not. (B) Table showing the performance of the 6 models in PlastNN. Each model was trained on five-sixths of the training set and cross-validated on the remaining one-sixth. Values shown are accuracy, sensitivity, specificity, NPV, and PPV on the cross-validation set. The final values reported are the average and standard deviation over all 6 models. (C) Comparison of accuracy, sensitivity, specificity, NPV, and PPV for 3 previous algorithms and PlastNN. NPV, negative predictive value; PlastNN, Apicoplast Neural Network; PPV, positive predictive value.