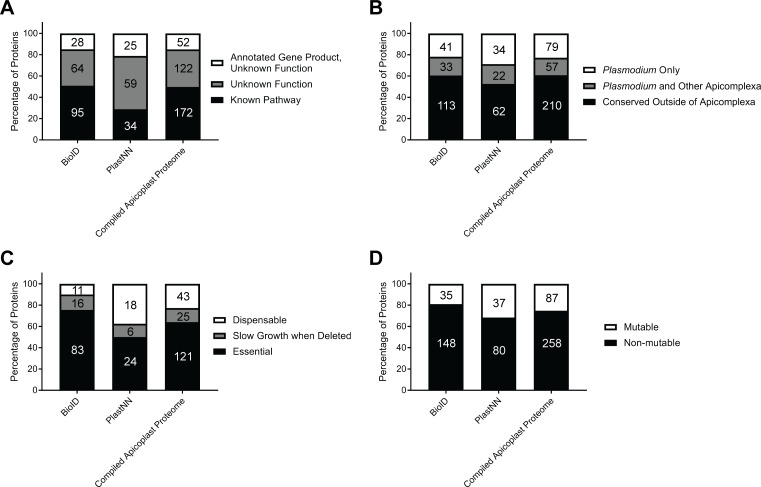
Fig 5. Apicoplast BioID identifies novel and essential proteins.
(A) Percentage of proteins identified that have 1) annotated gene products but unknown function, 2) gene products annotated explicitly with “unknown function,” or 3) annotated gene products and function in a known cellular pathway. (B) Percentage of proteins identified that are Plasmodium- or Apicomplexa-specific based on OrthoMCL-DB. (C) Percentage of proteins identified that are essential, cause slow growth when deleted, or are dispensable based on PlasmoGEM essentiality data of P. berghei orthologs [41]. (D) Percentage of proteins identified that were classified as mutable or nonmutable based on genome-scale transposon mutagenesis in P. falciparum [42]. In each panel, absolute numbers of proteins are indicated within bars. Tabulated data are available in S1 Data. BioID, proximity-dependent biotin identification; OrthoMCL-DB, Ortholog Groups of Protein Sequences database; PlasmoGEM, Plasmodium Genetic Modification project.