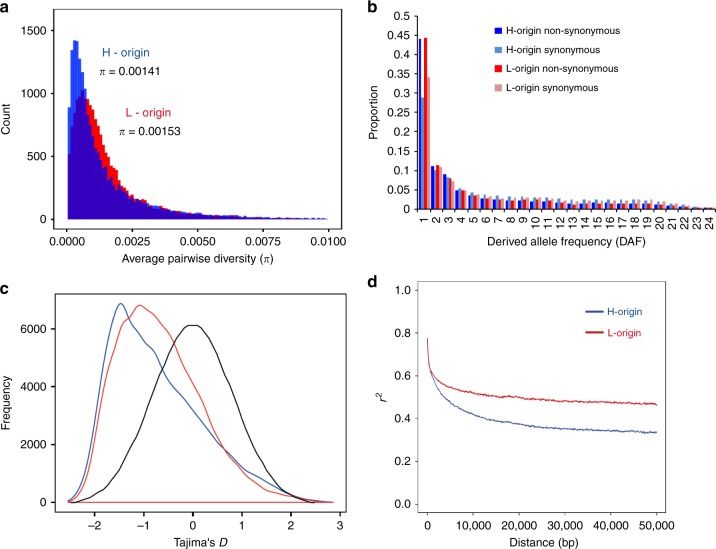
Fig. 1.
Genome-wide diversity and linkage disequilibrium. a Nucleotide diversity (average pairwise diversity, π) of halleri-origin (H-origin) and lyrata-origin (L-origin) coding sequences. b H-origin and L-origin genes show no significant differences in proportions of non-synonymous and synonymous substitutions (χ2-test, p-value = 0.58), and the majority of substitutions are at low frequency. c Tajima’s D distributions for both genomes (blue density curve = H-origin, red density curve = L-origin) show departures from neutrality (black density curve where neutral = 0); mean values for both distributions are negative (Table 1). d The mean decay of linkage disequilibrium (LD) estimated using 50 kb sliding windows shows mean LD decay <20 kb for both H-origin (blue) and L-origin (red) genomes